Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA
- PMID: 24167255
- PMCID: PMC3839702
- DOI: 10.1073/pnas.1310240110
Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA
Abstract
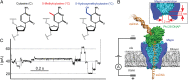
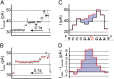
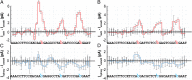
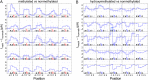
Precise and efficient mapping of epigenetic markers on DNA may become an important clinical tool for prediction and identification of ailments. Methylated CpG sites are involved in gene expression and are biomarkers for diseases such as cancer. Here, we use the engineered biological protein pore Mycobacterium smegmatis porin A (MspA) to detect and map 5-methylcytosine and 5-hydroxymethylcytosine within single strands of DNA. In this unique single-molecule tool, a phi29 DNA polymerase draws ssDNA through the pore in single-nucleotide steps, and the ion current through the pore is recorded. Comparing current levels generated with DNA containing methylated CpG sites to current levels obtained with unmethylated copies of the DNA reveals the precise location of methylated CpG sites. Hydroxymethylation is distinct from methylation and can also be mapped. With a single read, the detection efficiency in a quasirandom DNA strand is 97.5 ± 0.7% for methylation and 97 ± 0.9% for hydroxymethylation.
Keywords: DNA hydroxymethylation; DNA methylation; nanopore DNA sequencing; nanotechnology; next generation sequencing.
Conflict of interest statement
Conflict of interest statement: A.H.L., I.M.D., and J.H.G have filed a provisional patent on the detection strategy described herein. I.M.D. and J.H.G. have an interest with Illumina Inc. through licensed technology.
Figures


Comment in
-
Protein nanopores to detect DNA methylation.Nat Methods. 2014 Jan;11(1):8. doi: 10.1038/nmeth.2798. Nat Methods. 2014. PMID: 24524128 No abstract available.
References
-
- Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. - PubMed
-
- Marx V. Epigenetics: Reading the second genomic code. Nature. 2012;491(7422):143–147. - PubMed
-
- Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22(22):4632–4642. - PubMed
-
- Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: Modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

