Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography
- PMID: 24167270
- PMCID: PMC3832033
- DOI: 10.1073/pnas.1313156110
Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography
Abstract
Cryoelectron microscopy and X-ray crystallography have recently been used to generate structural models that likely represent the unliganded closed-channel conformation and the fully liganded open-channel conformation of different members of the nicotinic-receptor superfamily. To characterize the structure of the closed-channel conformation in its liganded state, we identified a number of positions in the loop between transmembrane segments 2 (M2) and 3 (M3) of a proton-gated ortholog from the bacterium Gloeobacter violaceus (GLIC) where mutations to alanine reduce the liganded-gating equilibrium constant, and solved the crystal structures of two such mutants (T25'A and Y27'A) at pH ~4.0. At the level of backbone atoms, the liganded closed-channel model presented here differs from the liganded open-channel structure of GLIC in the pre-M1 linker, the M3-M4 loop, and much more prominently, in the extracellular half of the pore lining, where the more pronounced tilt of the closed-channel M2 α-helices toward the pore's long axis narrows the permeation pathway. On the other hand, no differences between the liganded closed-channel and open-channel models could be detected at the level of the extracellular domain, where conformational changes are expected to underlie the low-to-high proton-affinity switch that drives gating of proton-bound channels. Thus, the liganded closed-channel model is nearly indistinguishable from the recently described "locally closed" structure. However, because cross-linking strategies (which could have stabilized unstable conformations) and mutations involving ionizable side chains (which could have affected proton-gated channel activation) were purposely avoided, we favor the notion that this structure represents one of the end states of liganded gating rather than an unstable intermediate.
Keywords: allostery; electrophysiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
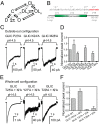

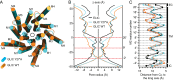
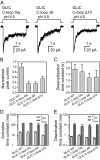
References
-
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346(4):967–989. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

