Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups
- PMID: 24167280
- PMCID: PMC3832011
- DOI: 10.1073/pnas.1318639110
Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups
Abstract
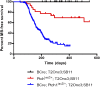
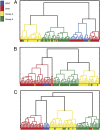
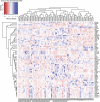
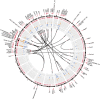
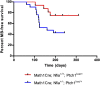
The Sleeping Beauty (SB) transposon mutagenesis screen is a powerful tool to facilitate the discovery of cancer genes that drive tumorigenesis in mouse models. In this study, we sought to identify genes that functionally cooperate with sonic hedgehog signaling to initiate medulloblastoma (MB), a tumor of the cerebellum. By combining SB mutagenesis with Patched1 heterozygous mice (Ptch1(lacZ/+)), we observed an increased frequency of MB and decreased tumor-free survival compared with Ptch1(lacZ/+) controls. From an analysis of 85 tumors, we identified 77 common insertion sites that map to 56 genes potentially driving increased tumorigenesis. The common insertion site genes identified in the mutagenesis screen were mapped to human orthologs, which were used to select probes and corresponding expression data from an independent set of previously described human MB samples, and surprisingly were capable of accurately clustering known molecular subgroups of MB, thereby defining common regulatory networks underlying all forms of MB irrespective of subgroup. We performed a network analysis to discover the likely mechanisms of action of subnetworks and used an in vivo model to confirm a role for a highly ranked candidate gene, Nfia, in promoting MB formation. Our analysis implicates candidate cancer genes in the deregulation of apoptosis and translational elongation, and reveals a strong signature of transcriptional regulation that will have broad impact on expression programs in MB. These networks provide functional insights into the complex biology of human MB and identify potential avenues for intervention common to all clinical subgroups.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gajjar A, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7(10):813–820. - PubMed
-
- Bhat SR, et al. Profile of daily life in children with brain tumors: An assessment of health-related quality of life. J Clin Oncol. 2005;23(24):5493–5500. - PubMed
-
- Palmer SL, Reddick WE, Gajjar A. Understanding the cognitive impact on children who are treated for medulloblastoma. J Pediatr Psychol. 2007;32(9):1040–1049. - PubMed
-
- Thompson MC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

