Isozyme-specific ligands for O-acetylserine sulfhydrylase, a novel antibiotic target
- PMID: 24167577
- PMCID: PMC3805590
- DOI: 10.1371/journal.pone.0077558
Isozyme-specific ligands for O-acetylserine sulfhydrylase, a novel antibiotic target
Abstract
The last step of cysteine biosynthesis in bacteria and plants is catalyzed by O-acetylserine sulfhydrylase. In bacteria, two isozymes, O-acetylserine sulfhydrylase-A and O-acetylserine sulfhydrylase-B, have been identified that share similar binding sites, although the respective specific functions are still debated. O-acetylserine sulfhydrylase plays a key role in the adaptation of bacteria to the host environment, in the defense mechanisms to oxidative stress and in antibiotic resistance. Because mammals synthesize cysteine from methionine and lack O-acetylserine sulfhydrylase, the enzyme is a potential target for antimicrobials. With this aim, we first identified potential inhibitors of the two isozymes via a ligand- and structure-based in silico screening of a subset of the ZINC library using FLAP. The binding affinities of the most promising candidates were measured in vitro on purified O-acetylserine sulfhydrylase-A and O-acetylserine sulfhydrylase-B from Salmonella typhimurium by a direct method that exploits the change in the cofactor fluorescence. Two molecules were identified with dissociation constants of 3.7 and 33 µM for O-acetylserine sulfhydrylase-A and O-acetylserine sulfhydrylase-B, respectively. Because GRID analysis of the two isoenzymes indicates the presence of a few common pharmacophoric features, cross binding titrations were carried out. It was found that the best binder for O-acetylserine sulfhydrylase-B exhibits a dissociation constant of 29 µM for O-acetylserine sulfhydrylase-A, thus displaying a limited selectivity, whereas the best binder for O-acetylserine sulfhydrylase-A exhibits a dissociation constant of 50 µM for O-acetylserine sulfhydrylase-B and is thus 8-fold selective towards the former isozyme. Therefore, isoform-specific and isoform-independent ligands allow to either selectively target the isozyme that predominantly supports bacteria during infection and long-term survival or to completely block bacterial cysteine biosynthesis.
Conflict of interest statement
Figures




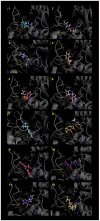
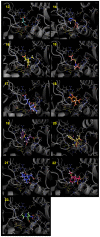

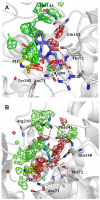

References
-
- Kessler D (2006) Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol Rev 30: 825–840. - PubMed
-
- Beinert H (2000) A tribute to sulfur. Eur J Biochem 267: 5657–5664. - PubMed
-
- Muller S, Liebau E, Walter RD, Krauth-Siegel RL (2003) Thiol-based redox metabolism of protozoan parasites. Trends Parasitol 19: 320–328. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

