Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut
- PMID: 24167633
- PMCID: PMC3805511
- DOI: 10.1371/journal.pone.0078555
Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut
Abstract
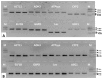
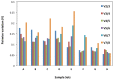
The quantitative real-time PCR (qPCR) based techniques have become essential for gene expression studies and high-throughput molecular characterization of transgenic events. Normalizing to reference gene in relative quantification make results from qPCR more reliable when compared to absolute quantification, but requires robust reference genes. Since, ideal reference gene should be species specific, no single internal control gene is universal for use as a reference gene across various plant developmental stages and diverse growth conditions. Here, we present validation studies of multiple stably expressed reference genes in cultivated peanut with minimal variations in temporal and spatial expression when subjected to various biotic and abiotic stresses. Stability in the expression of eight candidate reference genes including ADH3, ACT11, ATPsyn, CYP2, ELF1B, G6PD, LEC and UBC1 was compared in diverse peanut plant samples. The samples were categorized into distinct experimental sets to check the suitability of candidate genes for accurate and reliable normalization of gene expression using qPCR. Stability in expression of the references genes in eight sets of samples was determined by geNorm and NormFinder methods. While three candidate reference genes including ADH3, G6PD and ELF1B were identified to be stably expressed across experiments, LEC was observed to be the least stable, and hence must be avoided for gene expression studies in peanut. Inclusion of the former two genes gave sufficiently reliable results; nonetheless, the addition of the third reference gene ELF1B may be potentially better in a diverse set of tissue samples of peanut.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

