The role of Tal2 and Tal1 in the differentiation of midbrain GABAergic neuron precursors
- PMID: 24167708
- PMCID: PMC3798194
- DOI: 10.1242/bio.20135041
The role of Tal2 and Tal1 in the differentiation of midbrain GABAergic neuron precursors
Abstract
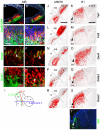
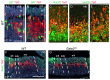
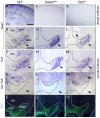
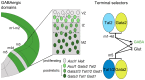
Midbrain- and hindbrain-derived GABAergic interneurons are critical for regulation of sleep, respiratory, sensory-motor and motivational processes, and they are implicated in human neurological disorders. However, the precise mechanisms that underlie generation of GABAergic neuron diversity in the midbrain-hindbrain region are poorly understood. Here, we show unique and overlapping requirements for the related bHLH proteins Tal1 and Tal2 in GABAergic neurogenesis in the midbrain. We show that Tal2 and Tal1 are specifically and sequentially activated during midbrain GABAergic neurogenesis. Similar to Gata2, a post-mitotic selector of the midbrain GABAergic neuron identity, Tal2 expression is activated very early during GABAergic neuron differentiation. Although the expression of Tal2 and Gata2 genes are independent of each other, Tal2 is important for normal midbrain GABAergic neurogenesis, possibly as a partner of Gata2. In the absence of Tal2, the majority of midbrain GABAergic neurons switch to a glutamatergic-like phenotype. In contrast, Tal1 expression is activated in a Gata2 and Tal2 dependent fashion in the more mature midbrain GABAergic neuron precursors, but Tal1 alone is not required for GABAergic neuron differentiation from the midbrain neuroepithelium. However, inactivation of both Tal2 and Tal1 in the developing midbrain suggests that the two factors co-operate to guide GABAergic neuron differentiation in a specific ventro-lateral midbrain domain. The observed similarities and differences between Tal1/Tal2 and Gata2 mutants suggest both co-operative and unique roles for these factors in determination of midbrain GABAergic neuron identities.
Keywords: Brain development; Dopaminergic neuron; GABAergic neuron; Gata; Hindbrain; Midbrain; Mouse; Neurogenesis; Rhombomere 1; Scl; Substantia nigra pars reticulata (SNpr); Tal; Transcription factor; Ventral tegmental area (VTA).
Conflict of interest statement
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

