Pulsed EPR study of amino acid and tetrahydropterin binding in a tyrosine hydroxylase nitric oxide complex: evidence for substrate rearrangements in the formation of the oxygen-reactive complex
- PMID: 24168553
- PMCID: PMC3902855
- DOI: 10.1021/bi4010914
Pulsed EPR study of amino acid and tetrahydropterin binding in a tyrosine hydroxylase nitric oxide complex: evidence for substrate rearrangements in the formation of the oxygen-reactive complex
Abstract
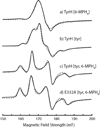
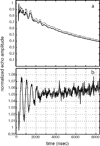
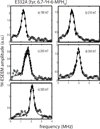
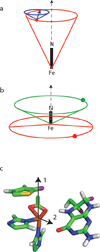
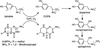
Tyrosine hydroxylase is a nonheme iron enzyme found in the nervous system that catalyzes the hydroxylation of tyrosine to form l-3,4-dihydroxyphenylalanine, the rate-limiting step in the biosynthesis of the catecholamine neurotransmitters. Catalysis requires the binding of three substrates: tyrosine, tetrahydrobiopterin, and molecular oxygen. We have used nitric oxide as an O₂ surrogate to poise Fe(II) at the catalytic site in an S = 3/2, {FeNO}⁷ form amenable to EPR spectroscopy. ²H-electron spin echo envelope modulation was then used to measure the distance and orientation of specifically deuterated substrate tyrosine and cofactor 6-methyltetrahydropterin with respect to the magnetic axes of the {FeNO}⁷ paramagnetic center. Our results show that the addition of tyrosine triggers a conformational change in the enzyme that reduces the distance from the {FeNO}⁷ center to the closest deuteron on 6,7-²H-6-methyltetrahydropterin from >5.9 Å to 4.4 ± 0.2 Å. Conversely, the addition of 6-methyltetrahydropterin to enzyme samples treated with 3,5-²H-tyrosine resulted in reorientation of the magnetic axes of the S = 3/2, {FeNO}⁷ center with respect to the deuterated substrate. Taken together, these results show that the coordination of both substrate and cofactor direct the coordination of NO to Fe(II) at the active site. Parallel studies of a quaternary complex of an uncoupled tyrosine hydroxylase variant, E332A, show no change in the hyperfine coupling to substrate tyrosine and cofactor 6-methyltetrahydropterin. Our results are discussed in the context of previous spectroscopic and X-ray crystallographic studies done on tyrosine hydroxylase and phenylalanine hydroxylase.
Figures





References
-
- Fitzpatrick PF. Tyrosine Hydroxylase. Annu. Rev. Biochem. 1999;68:355–381. - PubMed
-
- Ludecke B, Knappskog PM, Clayton PT, Surtees RAH, Clelland JD, Heales SJR, Brand MP, Batholome K, Flatmark T. Recessively inherited L-DOPA responsive Parkinsonism in infancy caused by point mutation of the tyrosine hydroxylase gene. Hum. Mol. Genet. 1996;5:1023–1028. - PubMed
-
- Ludecke B, Dworniczak B, Bartholome K. A point mutation in the tyrosine hydroxylase gene associated with Segawa's Syndrome. Hum. Genet. 1995;95:123–125. - PubMed
-
- Smyth C, Kalsi G, Brynjolfsson J, O'Neill J, Curtis D, Rifkin L, Maloney E, Murphy P, Sherrington R, Petursson H, Gurling H. Further tests for linkage of bipolar affective disorder to the tyrosine hydroxylase gene locus. Am. J. Psychiatry. 1996;153:271–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

