Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes
- PMID: 24169304
- PMCID: PMC3962528
- DOI: 10.4161/epi.26792
Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes
Abstract
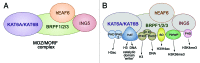
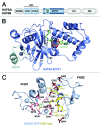
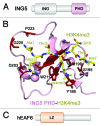
The MOZ/MORF complexes represent an example of a chromatin-binding assembly whose recruitment to specific genomic regions and activity can be fine-tuned by posttranslational modifications of histones. Here we detail the structures and biological functions of epigenetic readers present in the four core subunits of the MOZ/MORF complexes, highlight the imperative role of combinatorial readout by the multiple readers, and discuss new research directions to advance our understanding of histone acetylation.
Keywords: BRPF; ING5; KAT6A; KAT6B; MORF; MOZ; PHD; acetylation; histone; reader.
Figures

Similar articles
-
Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes.Mol Cell Biol. 2008 Nov;28(22):6828-43. doi: 10.1128/MCB.01297-08. Epub 2008 Sep 15. Mol Cell Biol. 2008. PMID: 18794358 Free PMC article.
-
Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin.J Mol Biol. 2012 Dec 14;424(5):328-38. doi: 10.1016/j.jmb.2012.10.004. Epub 2012 Oct 12. J Mol Biol. 2012. PMID: 23063713 Free PMC article.
-
Recognition of unmodified histone H3 by the first PHD finger of bromodomain-PHD finger protein 2 provides insights into the regulation of histone acetyltransferases monocytic leukemic zinc-finger protein (MOZ) and MOZ-related factor (MORF).J Biol Chem. 2011 Oct 21;286(42):36944-55. doi: 10.1074/jbc.M111.244400. Epub 2011 Aug 31. J Biol Chem. 2011. PMID: 21880731 Free PMC article.
-
The MOZ histone acetyltransferase in epigenetic signaling and disease.J Cell Physiol. 2014 Nov;229(11):1571-4. doi: 10.1002/jcp.24617. J Cell Physiol. 2014. PMID: 24633655 Free PMC article. Review.
-
MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease.Biochim Biophys Acta. 2015 Aug;1853(8):1818-26. doi: 10.1016/j.bbamcr.2015.04.014. Epub 2015 Apr 25. Biochim Biophys Acta. 2015. PMID: 25920810 Review.
Cited by
-
Histone-binding of DPF2 mediates its repressive role in myeloid differentiation.Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):6016-6021. doi: 10.1073/pnas.1700328114. Epub 2017 May 22. Proc Natl Acad Sci U S A. 2017. PMID: 28533407 Free PMC article.
-
Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2.Nucleic Acids Res. 2017 Jun 2;45(10):5707-5719. doi: 10.1093/nar/gkx142. Nucleic Acids Res. 2017. PMID: 28334966 Free PMC article.
-
The winged helix domain of MORF binds CpG islands and the TAZ2 domain of p300.iScience. 2024 Feb 29;27(4):109367. doi: 10.1016/j.isci.2024.109367. eCollection 2024 Apr 19. iScience. 2024. PMID: 38500836 Free PMC article.
-
Next-Generation Drugs and Probes for Chromatin Biology: From Targeted Protein Degradation to Phase Separation.Molecules. 2018 Aug 6;23(8):1958. doi: 10.3390/molecules23081958. Molecules. 2018. PMID: 30082609 Free PMC article. Review.
-
Bromodomain-containing protein BRPF1 is a therapeutic target for liver cancer.Commun Biol. 2021 Jul 20;4(1):888. doi: 10.1038/s42003-021-02405-6. Commun Biol. 2021. PMID: 34285329 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
