Promoters maintain their relative activity levels under different growth conditions
- PMID: 24169404
- PMCID: PMC3817408
- DOI: 10.1038/msb.2013.59
Promoters maintain their relative activity levels under different growth conditions
Abstract
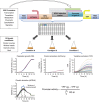
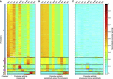
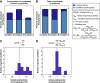
Most genes change expression levels across conditions, but it is unclear which of these changes represents specific regulation and what determines their quantitative degree. Here, we accurately measured activities of ~900 S. cerevisiae and ~1800 E. coli promoters using fluorescent reporters. We show that in both organisms 60-90% of promoters change their expression between conditions by a constant global scaling factor that depends only on the conditions and not on the promoter's identity. Quantifying such global effects allows precise characterization of specific regulation-promoters deviating from the global scale line. These are organized into few functionally related groups that also adhere to scale lines and preserve their relative activities across conditions. Thus, only several scaling factors suffice to accurately describe genome-wide expression profiles across conditions. We present a parameter-free passive resource allocation model that quantitatively accounts for the global scaling factors. It suggests that many changes in expression across conditions result from global effects and not specific regulation, and provides means for quantitative interpretation of expression profiles.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Saccharomyces cerevisiae JEN1 promoter activity is inversely related to concentration of repressing sugar.Appl Environ Microbiol. 2004 Jan;70(1):8-17. doi: 10.1128/AEM.70.1.8-17.2004. Appl Environ Microbiol. 2004. PMID: 14711620 Free PMC article.
-
Invariant distribution of promoter activities in Escherichia coli.PLoS Comput Biol. 2009 Oct;5(10):e1000545. doi: 10.1371/journal.pcbi.1000545. Epub 2009 Oct 23. PLoS Comput Biol. 2009. PMID: 19851443 Free PMC article.
-
Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector.J Bacteriol. 2004 Nov;186(21):7112-22. doi: 10.1128/JB.186.21.7112-7122.2004. J Bacteriol. 2004. PMID: 15489422 Free PMC article.
-
Thiol-based H2O2 signalling in microbial systems.Redox Biol. 2014 Feb 3;2:395-9. doi: 10.1016/j.redox.2014.01.015. eCollection 2014. Redox Biol. 2014. PMID: 24563858 Free PMC article. Review.
-
[Base excision repair: AP endonucleases and DNA polymerases].Genetika. 2005 Oct;41(10):1301-9. Genetika. 2005. PMID: 16316001 Review. Russian.
Cited by
-
Mechanistic links between cellular trade-offs, gene expression, and growth.Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):E1038-47. doi: 10.1073/pnas.1416533112. Epub 2015 Feb 18. Proc Natl Acad Sci U S A. 2015. PMID: 25695966 Free PMC article.
-
A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae.Microb Cell Fact. 2021 Oct 18;20(1):202. doi: 10.1186/s12934-021-01691-3. Microb Cell Fact. 2021. PMID: 34663323 Free PMC article.
-
Advancing biotechnology with CRISPR/Cas9: recent applications and patent landscape.J Ind Microbiol Biotechnol. 2018 Jul;45(7):467-480. doi: 10.1007/s10295-017-2000-6. Epub 2018 Jan 24. J Ind Microbiol Biotechnol. 2018. PMID: 29362972 Review.
-
The Biosynthetic Basis of Cell Size Control.Trends Cell Biol. 2015 Dec;25(12):793-802. doi: 10.1016/j.tcb.2015.10.006. Epub 2015 Nov 10. Trends Cell Biol. 2015. PMID: 26573465 Free PMC article. Review.
-
Global relationships in fluctuation and response in adaptive evolution.J R Soc Interface. 2015 Aug 6;12(109):20150482. doi: 10.1098/rsif.2015.0482. J R Soc Interface. 2015. PMID: 26202686 Free PMC article.
References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, Gebbia M, Talukder S, Yang A, Mnaimneh S, Terterov D, Coburn D, Li Yeo A, Yeo ZX, Clarke ND, Lieb JD et al. (2008) A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 - PMC - PubMed
-
- Bakel H Van, Holstege FCP (2008) A tutorial for DNA microarray expression profiling. Cell Evaluating: 22–28 , Available at: http://download.cell.com/images/edimages/etbr/VANBAKEL.PDF
-
- Balázsi G, Oltvai ZN (2007) A pitfall in series of microarrays: the position of probes affects the cross-correlation of gene expression profiles. Methods Mol Biol (Clifton, NJ) 377: 153–162 - PubMed
-
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, Cunningham ML, Deng S, Dressman HK, Fannin RD, Farin FM, Freedman JH, Fry RC, Harper A, Humble MC, Hurban P et al. (2005) Standardizing global gene expression analysis between laboratories and across platforms. Nat Meth 2: 351–356 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

