Elucidating novel hepatitis C virus-host interactions using combined mass spectrometry and functional genomics approaches
- PMID: 24169621
- PMCID: PMC3879614
- DOI: 10.1074/mcp.M113.030155
Elucidating novel hepatitis C virus-host interactions using combined mass spectrometry and functional genomics approaches
Abstract
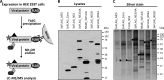
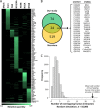
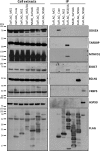
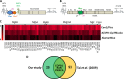
More than 170 million people worldwide are infected with the hepatitis C virus (HCV), for which future therapies are expected to rely upon a combination of oral antivirals. For a rapidly evolving virus like HCV, host-targeting antivirals are an attractive option. To decipher the role of novel HCV-host interactions, we used a proteomics approach combining immunoprecipitation of viral-host protein complexes coupled to mass spectrometry identification and functional genomics RNA interference screening of HCV partners. Here, we report the proteomics analyses of protein complexes associated with Core, NS2, NS3/4A, NS4B, NS5A, and NS5B proteins. We identified a stringent set of 98 human proteins interacting specifically with one of the viral proteins. The overlap with previous virus-host interaction studies demonstrates 24.5% shared HCV interactors overall (24/98), illustrating the reliability of the approach. The identified human proteins show enriched Gene Ontology terms associated with the endoplasmic reticulum, transport proteins with a major contribution of NS3/4A interactors, and transmembrane proteins for Core interactors. The interaction network emphasizes a high degree distribution, a high betweenness distribution, and high interconnectivity of targeted human proteins, in agreement with previous virus-host interactome studies. The set of HCV interactors also shows extensive enrichment for known targets of other viruses. The combined proteomic and gene silencing study revealed strong enrichment in modulators of HCV RNA replication, with the identification of 11 novel cofactors among our set of specific HCV partners. Finally, we report a novel immune evasion mechanism of NS3/4A protein based on its ability to affect nucleocytoplasmic transport of type I interferon-mediated signal transducer and activator of transcription 1 nuclear translocation. The study revealed highly stringent association between HCV interactors and their functional contribution to the viral replication cycle and pathogenesis.
Conflict of interest statement
Conflict of interest: D.L. is a shareholder in ViroCura Therapeutics Inc.
Figures





Similar articles
-
Landscape of protein-protein interactions during hepatitis C virus assembly and release.Microbiol Spectr. 2024 Feb 6;12(2):e0256222. doi: 10.1128/spectrum.02562-22. Epub 2024 Jan 17. Microbiol Spectr. 2024. PMID: 38230952 Free PMC article.
-
A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production.J Virol. 2013 Nov;87(21):11704-20. doi: 10.1128/JVI.01474-13. Epub 2013 Aug 28. J Virol. 2013. PMID: 23986595 Free PMC article.
-
Palmitoylation of Hepatitis C Virus NS2 Regulates Its Subcellular Localization and NS2-NS3 Autocleavage.J Virol. 2019 Dec 12;94(1):e00906-19. doi: 10.1128/JVI.00906-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597774 Free PMC article.
-
Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review.
-
The history of hepatitis C virus (HCV): Basic research reveals unique features in phylogeny, evolution and the viral life cycle with new perspectives for epidemic control.J Hepatol. 2016 Oct;65(1 Suppl):S2-S21. doi: 10.1016/j.jhep.2016.07.035. J Hepatol. 2016. PMID: 27641985 Review.
Cited by
-
Network-Guided Discovery of Influenza Virus Replication Host Factors.mBio. 2018 Dec 18;9(6):e02002-18. doi: 10.1128/mBio.02002-18. mBio. 2018. PMID: 30563907 Free PMC article.
-
Defining Pharmacological Targets by Analysis of Virus-Host Protein Interactions.Adv Protein Chem Struct Biol. 2018;111:223-242. doi: 10.1016/bs.apcsb.2017.11.001. Epub 2017 Dec 18. Adv Protein Chem Struct Biol. 2018. PMID: 29459033 Free PMC article. Review.
-
Metabolic modeling elucidates phenformin and atpenin A5 as broad-spectrum antiviral drugs against RNA viruses.Commun Biol. 2025 May 23;8(1):791. doi: 10.1038/s42003-025-08148-y. Commun Biol. 2025. PMID: 40410544 Free PMC article.
-
The lysine methyltransferase SMYD3 interacts with hepatitis C virus NS5A and is a negative regulator of viral particle production.Virology. 2014 Aug;462-463(100):34-41. doi: 10.1016/j.virol.2014.05.016. Epub 2014 Jun 14. Virology. 2014. PMID: 25092459 Free PMC article.
-
Hepatitis C Virus Infection and Intrinsic Disorder in the Signaling Pathways Induced by Toll-Like Receptors.Biology (Basel). 2022 Jul 21;11(7):1091. doi: 10.3390/biology11071091. Biology (Basel). 2022. PMID: 36101469 Free PMC article. Review.
References
-
- Poynard T., Yuen M. F., Ratziu V., Lai C. L. (2003) Viral hepatitis C. Lancet 362, 2095–2100 - PubMed
-
- Welsch C., Jesudian A., Zeuzem S., Jacobson I. (2012) New direct-acting antiviral agents for the treatment of hepatitis C virus infection and perspectives. Gut 61 Suppl 1, i36–i46 - PubMed
-
- Romero-Brey I., Merz A., Chiramel A., Lee J. Y., Chlanda P., Haselman U., Santarella-Mellwig R., Habermann A., Hoppe S., Kallis S., Walther P., Antony C., Krijnse-Locker J., Bartenschlager R. (2012) Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8, e1003056. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

