A fluorescence-based assay suitable for quantitative analysis of deadenylase enzyme activity
- PMID: 24170810
- PMCID: PMC3950723
- DOI: 10.1093/nar/gkt972
A fluorescence-based assay suitable for quantitative analysis of deadenylase enzyme activity
Abstract
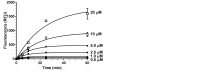
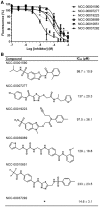
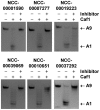
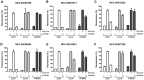
In eukaryotic cells, the shortening and removal of the poly(A) tail of cytoplasmic mRNA by deadenylase enzymes is a critical step in post-transcriptional gene regulation. The ribonuclease activity of deadenylase enzymes is attributed to either a DEDD (Asp-Glu-Asp-Asp) or an endonuclease-exonuclease-phosphatase domain. Both domains require the presence of two Mg2+ ions in the active site. To facilitate the biochemical analysis of deadenylase enzymes, we have developed a fluorescence-based deadenylase assay. The assay is based on end-point measurement, suitable for quantitative analysis and can be adapted for 96- and 384-well microplate formats. We demonstrate the utility of the assay by screening a chemical compound library, resulting in the identification of non-nucleoside inhibitors of the Caf1/CNOT7 enzyme, a catalytic subunit of the Ccr4-Not deadenylase complex. These compounds may be useful tools for the biochemical analysis of the Caf1/CNOT7 deadenylase subunit of the Ccr4-Not complex and indicate the feasibility of developing selective inhibitors of deadenylase enzymes using the fluorescence-based assay.
Figures


References
-
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. - PubMed
-
- Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol Cell Biol. 2008;9:337–344. - PubMed
-
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. - PubMed
-
- Wahle E, Winkler GS. RNA decay machines: Deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta. 2013;1829:561–570. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

