Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration
- PMID: 24170860
- PMCID: PMC3839752
- DOI: 10.1073/pnas.1318835110
Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration
Abstract
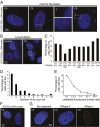
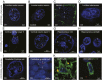
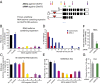
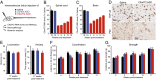
Expanded hexanucleotide repeats in the chromosome 9 open reading frame 72 (C9orf72) gene are the most common genetic cause of ALS and frontotemporal degeneration (FTD). Here, we identify nuclear RNA foci containing the hexanucleotide expansion (GGGGCC) in patient cells, including white blood cells, fibroblasts, glia, and multiple neuronal cell types (spinal motor, cortical, hippocampal, and cerebellar neurons). RNA foci are not present in sporadic ALS, familial ALS/FTD caused by other mutations (SOD1, TDP-43, or tau), Parkinson disease, or nonneurological controls. Antisense oligonucleotides (ASOs) are identified that reduce GGGGCC-containing nuclear foci without altering overall C9orf72 RNA levels. By contrast, siRNAs fail to reduce nuclear RNA foci despite marked reduction in overall C9orf72 RNAs. Sustained ASO-mediated lowering of C9orf72 RNAs throughout the CNS of mice is demonstrated to be well tolerated, producing no behavioral or pathological features characteristic of ALS/FTD and only limited RNA expression alterations. Genome-wide RNA profiling identifies an RNA signature in fibroblasts from patients with C9orf72 expansion. ASOs targeting sense strand repeat-containing RNAs do not correct this signature, a failure that may be explained, at least in part, by discovery of abundant RNA foci with C9orf72 repeats transcribed in the antisense (GGCCCC) direction, which are not affected by sense strand-targeting ASOs. Taken together, these findings support a therapeutic approach by ASO administration to reduce hexanucleotide repeat-containing RNAs and raise the potential importance of targeting expanded RNAs transcribed in both directions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Neurodegenerative disease: C9orf72 RNA foci--a therapeutic target for ALS and FTD?Nat Rev Neurol. 2013 Dec;9(12):659. doi: 10.1038/nrneurol.2013.244. Epub 2013 Nov 26. Nat Rev Neurol. 2013. PMID: 24275931 No abstract available.
References
-
- Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289(5485):1769–1773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

