Engineered proteins detect spontaneous DNA breakage in human and bacterial cells
- PMID: 24171103
- PMCID: PMC3809393
- DOI: 10.7554/eLife.01222
Engineered proteins detect spontaneous DNA breakage in human and bacterial cells
Abstract
Spontaneous DNA breaks instigate genomic changes that fuel cancer and evolution, yet direct quantification of double-strand breaks (DSBs) has been limited. Predominant sources of spontaneous DSBs remain elusive. We report synthetic technology for quantifying DSBs using fluorescent-protein fusions of double-strand DNA end-binding protein, Gam of bacteriophage Mu. In Escherichia coli GamGFP forms foci at chromosomal DSBs and pinpoints their subgenomic locations. Spontaneous DSBs occur mostly one per cell, and correspond with generations, supporting replicative models for spontaneous breakage, and providing the first true breakage rates. In mammalian cells GamGFP-labels laser-induced DSBs antagonized by end-binding protein Ku; co-localizes incompletely with DSB marker 53BP1 suggesting superior DSB-specificity; blocks resection; and demonstrates DNA breakage via APOBEC3A cytosine deaminase. We demonstrate directly that some spontaneous DSBs occur outside of S phase. The data illuminate spontaneous DNA breakage in E. coli and human cells and illustrate the versatility of fluorescent-Gam for interrogation of DSBs in living cells. DOI:http://dx.doi.org/10.7554/eLife.01222.001.
Keywords: DNA double-strand breaks; E. coli; GFP; Human; Mouse; endogenous DNA damage; fluorescent-protein fusions; spontaneous DNA breaks; synthetic biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
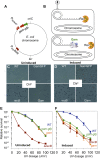
 ), recB− (
), recB− ( ), WT GamGFP, (
), WT GamGFP, ( ); WT Gam, (
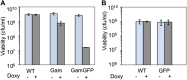
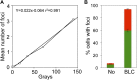
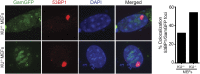
); WT Gam, ( ). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results. DOI:
). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results. DOI:
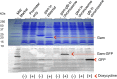

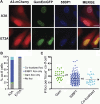
 ) and GamGFP foci (
) and GamGFP foci ( ) results in a yellow focus (
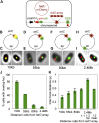
) results in a yellow focus ( ). (C, E, G, I) Representative fluorescence microscopy results show co-localization of mCherry and GamGFP (yellow foci) at 10 kb (C), and non-overlapping foci at 55 kb (E), 80 kb (G), 2.4 Mb (I) in strains SMR16600, SMR16711, SMR16713, and SMR16606. (J) Percentage of cells with yellow overlapped foci at each distance. (K) Mean interfocal distances. At 2.4 Mb, there were frequently two red foci per one green focus, reflecting more copies of ori- than ter-proximal DNA during replication. The greater interfocal distance (far) is plotted separately from the shorter (near), and cells with 1:1 ratios were counted separately. Data represent three independent experiments, error bars indicate SEM, with the number of cells counted in all three totaling: 298, 10 kb; 204, 55 kb; 333, 80 kb; and 1347, 2.4 Mb. DOI:
). (C, E, G, I) Representative fluorescence microscopy results show co-localization of mCherry and GamGFP (yellow foci) at 10 kb (C), and non-overlapping foci at 55 kb (E), 80 kb (G), 2.4 Mb (I) in strains SMR16600, SMR16711, SMR16713, and SMR16606. (J) Percentage of cells with yellow overlapped foci at each distance. (K) Mean interfocal distances. At 2.4 Mb, there were frequently two red foci per one green focus, reflecting more copies of ori- than ter-proximal DNA during replication. The greater interfocal distance (far) is plotted separately from the shorter (near), and cells with 1:1 ratios were counted separately. Data represent three independent experiments, error bars indicate SEM, with the number of cells counted in all three totaling: 298, 10 kb; 204, 55 kb; 333, 80 kb; and 1347, 2.4 Mb. DOI:
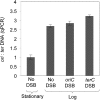
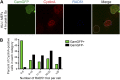
 ), number of cell divisions; green (
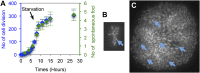
), number of cell divisions; green ( ), cumulative number of spontaneous foci that appear in each microfluidics micro-colony (mean ± SEM, six microcolonies). (B) Representative 2-hr micro-colony with a GamGFP focus (arrow). (C) Representative 15-hr micro-colony with GamGFP foci (arrows). DOI:
), cumulative number of spontaneous foci that appear in each microfluidics micro-colony (mean ± SEM, six microcolonies). (B) Representative 2-hr micro-colony with a GamGFP focus (arrow). (C) Representative 15-hr micro-colony with GamGFP foci (arrows). DOI:



Comment in
-
Proteins pinpoint double strand breaks.Elife. 2013 Oct 29;2:e01561. doi: 10.7554/eLife.01561. Elife. 2013. PMID: 24171106 Free PMC article.
Similar articles
-
Proteins pinpoint double strand breaks.Elife. 2013 Oct 29;2:e01561. doi: 10.7554/eLife.01561. Elife. 2013. PMID: 24171106 Free PMC article.
-
Fluorescent fusions of the N protein of phage Mu label DNA damage in living cells.DNA Repair (Amst). 2018 Dec;72:86-92. doi: 10.1016/j.dnarep.2018.09.005. Epub 2018 Sep 14. DNA Repair (Amst). 2018. PMID: 30268364 Free PMC article.
-
Two- and three-dimensional live cell imaging of DNA damage response proteins.J Vis Exp. 2012 Sep 28;(67):4251. doi: 10.3791/4251. J Vis Exp. 2012. PMID: 23052275 Free PMC article.
-
The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
-
Biochemical mechanism of DSB end resection and its regulation.DNA Repair (Amst). 2015 Aug;32:66-74. doi: 10.1016/j.dnarep.2015.04.015. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956866 Free PMC article. Review.
Cited by
-
Life, the genome and everything.J Bacteriol. 2023 Dec 19;205(12):e0027223. doi: 10.1128/jb.00272-23. Epub 2023 Nov 29. J Bacteriol. 2023. PMID: 38018999 Free PMC article.
-
MioC and GidA proteins promote cell division in E. coli.Front Microbiol. 2015 May 28;6:516. doi: 10.3389/fmicb.2015.00516. eCollection 2015. Front Microbiol. 2015. PMID: 26074904 Free PMC article.
-
Regulation of the error-prone DNA polymerase Polκ by oncogenic signaling and its contribution to drug resistance.Sci Signal. 2020 Apr 28;13(629):eaau1453. doi: 10.1126/scisignal.aau1453. Sci Signal. 2020. PMID: 32345725 Free PMC article.
-
Biased Gene Conversion in Rhizobium etli Is Caused by Preferential Double-Strand Breaks on One of the Recombining Homologs.J Bacteriol. 2015 Nov 23;198(3):591-9. doi: 10.1128/JB.00768-15. Print 2016 Feb 1. J Bacteriol. 2015. PMID: 26598365 Free PMC article.
-
CRISPR-Cas adaptation: insights into the mechanism of action.Nat Rev Microbiol. 2016 Feb;14(2):67-76. doi: 10.1038/nrmicro.2015.14. Epub 2016 Jan 11. Nat Rev Microbiol. 2016. PMID: 26751509 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- DP1-CA174424/CA/NCI NIH HHS/United States
- R01-GM88653/GM/NIGMS NIH HHS/United States
- R01 GM102679/GM/NIGMS NIH HHS/United States
- P01-GM091743/GM/NIGMS NIH HHS/United States
- R01-AI064046/AI/NIAID NIH HHS/United States
- CA097175/CA/NCI NIH HHS/United States
- F32-GM095267/GM/NIGMS NIH HHS/United States
- R01 GM088653/GM/NIGMS NIH HHS/United States
- R01 GM053158/GM/NIGMS NIH HHS/United States
- R01-GM53158/GM/NIGMS NIH HHS/United States
- DP1 CA174424/CA/NCI NIH HHS/United States
- CA127945/CA/NCI NIH HHS/United States
- R01-GM102679/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

