The effect of genomic information on optimal contribution selection in livestock breeding programs
- PMID: 24171942
- PMCID: PMC4176995
- DOI: 10.1186/1297-9686-45-44
The effect of genomic information on optimal contribution selection in livestock breeding programs
Abstract
Background: Long-term benefits in animal breeding programs require that increases in genetic merit be balanced with the need to maintain diversity (lost due to inbreeding). This can be achieved by using optimal contribution selection. The availability of high-density DNA marker information enables the incorporation of genomic data into optimal contribution selection but this raises the question about how this information affects the balance between genetic merit and diversity.
Methods: The effect of using genomic information in optimal contribution selection was examined based on simulated and real data on dairy bulls. We compared the genetic merit of selected animals at various levels of co-ancestry restrictions when using estimated breeding values based on parent average, genomic or progeny test information. Furthermore, we estimated the proportion of variation in estimated breeding values that is due to within-family differences.
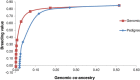
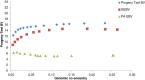
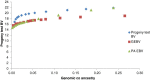
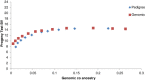
Results: Optimal selection on genomic estimated breeding values increased genetic gain. Genetic merit was further increased using genomic rather than pedigree-based measures of co-ancestry under an inbreeding restriction policy. Using genomic instead of pedigree relationships to restrict inbreeding had a significant effect only when the population consisted of many large full-sib families; with a half-sib family structure, no difference was observed. In real data from dairy bulls, optimal contribution selection based on genomic estimated breeding values allowed for additional improvements in genetic merit at low to moderate inbreeding levels. Genomic estimated breeding values were more accurate and showed more within-family variation than parent average breeding values; for genomic estimated breeding values, 30 to 40% of the variation was due to within-family differences. Finally, there was no difference between constraining inbreeding via pedigree or genomic relationships in the real data.
Conclusions: The use of genomic estimated breeding values increased genetic gain in optimal contribution selection. Genomic estimated breeding values were more accurate and showed more within-family variation, which led to higher genetic gains for the same restriction on inbreeding. Using genomic relationships to restrict inbreeding provided no additional gain, except in the case of very large full-sib families.
Figures



Similar articles
-
Effects of genomic selection on genetic improvement, inbreeding, and merit of young versus proven bulls.J Dairy Sci. 2011 Mar;94(3):1559-67. doi: 10.3168/jds.2010-3354. J Dairy Sci. 2011. PMID: 21338821
-
Genotyping more cows increases genetic gain and reduces rate of true inbreeding in a dairy cattle breeding scheme using female reproductive technologies.J Dairy Sci. 2020 Jan;103(1):597-606. doi: 10.3168/jds.2019-16974. Epub 2019 Nov 14. J Dairy Sci. 2020. PMID: 31733861
-
Effects of germplasm exchange strategies on genetic gain, homozygosity, and genetic diversity in dairy stud populations: A simulation study.J Dairy Sci. 2024 Dec;107(12):11149-11163. doi: 10.3168/jds.2024-24992. Epub 2024 Aug 30. J Dairy Sci. 2024. PMID: 39216524
-
Invited review: Genomic selection in dairy cattle: progress and challenges.J Dairy Sci. 2009 Feb;92(2):433-43. doi: 10.3168/jds.2008-1646. J Dairy Sci. 2009. PMID: 19164653 Review.
-
Integrating genomic selection into dairy cattle breeding programmes: a review.Animal. 2013 May;7(5):705-13. doi: 10.1017/S1751731112002248. Epub 2012 Dec 3. Animal. 2013. PMID: 23200196 Review.
Cited by
-
Optimized breeding strategies to harness genetic resources with different performance levels.BMC Genomics. 2020 May 11;21(1):349. doi: 10.1186/s12864-020-6756-0. BMC Genomics. 2020. PMID: 32393177 Free PMC article.
-
Pedigree relationships to control inbreeding in optimum-contribution selection realise more genetic gain than genomic relationships.Genet Sel Evol. 2019 Jul 8;51(1):39. doi: 10.1186/s12711-019-0475-5. Genet Sel Evol. 2019. PMID: 31286868 Free PMC article.
-
Evaluation of Linear Programming and Optimal Contribution Selection Approaches for Long-Term Selection on Beef Cattle Breeding.Biology (Basel). 2023 Aug 23;12(9):1157. doi: 10.3390/biology12091157. Biology (Basel). 2023. PMID: 37759557 Free PMC article.
-
Impact of kinship matrices on genetic gain and inbreeding with optimum contribution selection in a genomic dairy cattle breeding program.Genet Sel Evol. 2023 Jul 17;55(1):48. doi: 10.1186/s12711-023-00826-x. Genet Sel Evol. 2023. PMID: 37460999 Free PMC article.
-
Stacked neural network for predicting polygenic risk score.Sci Rep. 2024 May 21;14(1):11632. doi: 10.1038/s41598-024-62513-1. Sci Rep. 2024. PMID: 38773257 Free PMC article.
References
-
- Falconer DS, MacKay TFC. Introduction to quantitative genetics. 4. Harlow: Longman Scientific & Technical; 1996.
-
- Wray NR, Goddard ME. Increasing long-term response to selection. Genet Sel Evol. 1994;26:431–451. doi: 10.1186/1297-9686-26-5-431. - DOI
-
- Meuwissen THE. Maximizing the response of selection with a predetermined rate of inbreeding. J Anim Sci. 1997;75:934–940. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

