Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013
- PMID: 24172151
- PMCID: PMC3911414
- DOI: 10.1128/JCM.02680-13
Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013
Abstract
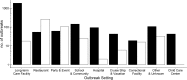
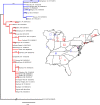
Noroviruses are the leading cause of epidemic acute gastroenteritis in the United States. From September 2009 through August 2013, 3,960 norovirus outbreaks were reported to CaliciNet. Of the 2,895 outbreaks with a known transmission route, person-to-person and food-borne transmissions were reported for 2,425 (83.7%) and 465 (16.1%) of the outbreaks, respectively. A total of 2,475 outbreaks (62.5%) occurred in long-term care facilities (LTCF), 389 (9.8%) in restaurants, and 227 (5.7%) in schools. A total of 435 outbreaks (11%) were typed as genogroup I (GI) and 3,525 (89%) as GII noroviruses. GII.4 viruses caused 2,853 (72%) of all outbreaks, of which 94% typed as either GII.4 New Orleans or GII.4 Sydney. In addition, three non-GII.4 viruses, i.e., GII.12, GII.1, and GI.6, caused 528 (13%) of all outbreaks. Several non-GII.4 genotypes (GI.3, GI.6, GI.7, GII.3, GII.6, and GII.12) were significantly more associated with food-borne transmission (odds ratio, 1.9 to 7.1; P < 0.05). Patients in LTCF and people ≥65 years of age were at higher risk for GII.4 infections than those in other settings and with other genotypes (P < 0.05). Phylogeographic analysis identified three major dispersions from two geographic locations that were responsible for the GI.6 outbreaks from 2011 to 2013. In conclusion, our data demonstrate the cyclic emergence of new (non-GII.4) norovirus strains, and several genotypes are more often associated with food-borne outbreaks. These surveillance data can be used to improve viral food-borne surveillance and to help guide studies to develop and evaluate targeted prevention methods such as norovirus vaccines, antivirals, and environmental decontamination methods.
Figures


References
-
- Payne DC, Vinjé J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT, Wikswo M, Shirley SH, Hall AJ, Lopman B, Parashar UD. 2013. Norovirus and medically attended gastroenteritis in U.S. children. N. Engl. J. Med. 368:1121–1130. 10.1056/NEJMsa1206589 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

