Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples
- PMID: 24172157
- PMCID: PMC3911411
- DOI: 10.1128/JCM.02452-13
Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples
Erratum in
- J Clin Microbiol. 2014 Aug;52(8):3136
Abstract
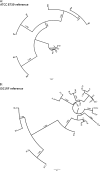
Whole-genome sequencing (WGS) is becoming available as a routine tool for clinical microbiology. If applied directly on clinical samples, this could further reduce diagnostic times and thereby improve control and treatment. A major bottleneck is the availability of fast and reliable bioinformatic tools. This study was conducted to evaluate the applicability of WGS directly on clinical samples and to develop easy-to-use bioinformatic tools for the analysis of sequencing data. Thirty-five random urine samples from patients with suspected urinary tract infections were examined using conventional microbiology, WGS of isolated bacteria, and direct sequencing on pellets from the urine samples. A rapid method for analyzing the sequence data was developed. Bacteria were cultivated from 19 samples but in pure cultures from only 17 samples. WGS improved the identification of the cultivated bacteria, and almost complete agreement was observed between phenotypic and predicted antimicrobial susceptibilities. Complete agreement was observed between species identification, multilocus sequence typing, and phylogenetic relationships for Escherichia coli and Enterococcus faecalis isolates when the results of WGS of cultured isolates and urine samples were directly compared. Sequencing directly from the urine enabled bacterial identification in polymicrobial samples. Additional putative pathogenic strains were observed in some culture-negative samples. WGS directly on clinical samples can provide clinically relevant information and drastically reduce diagnostic times. This may prove very useful, but the need for data analysis is still a hurdle to clinical implementation. To overcome this problem, a publicly available bioinformatic tool was developed in this study.
Figures
References
-
- Aarestrup FM, Brown EW, Detter C, Gerner-Smidt P, Gilmour MW, Harmsen D, Hendriksen RS, Hewson R, Heymann DL, Johansson K, Ijaz K, Keim PS, Koopmans M, Kroneman A, Lo Fo Wong D, Lund O, Palm D, Sawanpanyalert P, Sobel J, Schlundt J. 2012. Integrating genome-based informatics to modernize global disease monitoring, information sharing, and response. Emerg. Infect. Dis. 18:e1. 10.3201/eid/1811.120453 - DOI - PMC - PubMed
-
- Köser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 8:e1002824. 10.1371/journal.ppat.1002824 - DOI - PMC - PubMed
-
- Harris SR, Cartwright EJ, Török ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. 13:130–136. 10.1016/S1473-3099(12)70268-2 - DOI - PMC - PubMed
-
- Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2013. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 4:e00520–12. 10.1128/mBio.00520-12. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

