Molecular docking studies of marine diterpenes as inhibitors of wild-type and mutants HIV-1 reverse transcriptase
- PMID: 24172210
- PMCID: PMC3853719
- DOI: 10.3390/md11114127
Molecular docking studies of marine diterpenes as inhibitors of wild-type and mutants HIV-1 reverse transcriptase
Abstract
AIDS is a pandemic responsible for more than 35 million deaths. The emergence of resistant mutations due to drug use is the biggest cause of treatment failure. Marine organisms are sources of different molecules, some of which offer promising HIV-1 reverse transcriptase (RT) inhibitory activity, such as the diterpenes dolabelladienotriol (THD, IC50 = 16.5 µM), (6R)-6-hydroxydichotoma-3,14-diene-1,17-dial (HDD, IC50 = 10 µM) and (6R)-6-acetoxydichotoma-3,14-diene-1,17-dial (ADD, IC50 = 35 µM), isolated from a brown algae of the genus Dictyota, showing low toxicity. In this work, we evaluated the structure-activity relationship (SAR) of THD, HDD and ADD as anti HIV-1 RT, using a molecular modeling approach. The analyses of stereoelectronic parameters revealed a direct relationship between activity and HOMO (Highest Occupied Molecular Orbital)-LUMO (Lowest Unoccupied Molecular Orbital) gap (E(LUMO)-E(HOMO)), where antiviral profile increases with larger HOMO-LUMO gap values. We also performed molecular docking studies of THD into HIV-1 RT wild-type and 12 different mutants, which showed a seahorse conformation, hydrophobic interactions and hydrogen bonds with important residues of the binding pocket. Based on in vitro experiments and docking studies, we demonstrated that mutations have little influence in positioning and interactions of THD. Following a rational drug design, we suggest a modification of THD to improve its biological activity.
Figures
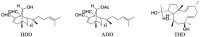



Similar articles
-
Antiviral activity of diterpenes isolated from the Brazilian marine alga Dictyota menstrualis against human immunodeficiency virus type 1 (HIV-1).Antiviral Res. 2004 Oct;64(1):69-76. doi: 10.1016/j.antiviral.2004.06.006. Antiviral Res. 2004. PMID: 15451181
-
Effects of diterpenes isolated from the Brazilian marine alga Dictyota menstrualis on HIV-1 reverse transcriptase.Planta Med. 2005 Nov;71(11):1019-24. doi: 10.1055/s-2005-873113. Planta Med. 2005. PMID: 16320202
-
Docking of non-nucleoside inhibitors: neotripterifordin and its derivatives to HIV-1 reverse transcriptase.Proteins. 2002 Dec 1;49(4):529-42. doi: 10.1002/prot.10233. Proteins. 2002. PMID: 12402361
-
Molecular Docking Studies of HIV-1 Resistance to Reverse Transcriptase Inhibitors: Mini-Review.Molecules. 2018 May 21;23(5):1233. doi: 10.3390/molecules23051233. Molecules. 2018. PMID: 29883406 Free PMC article. Review.
-
Recent progress in HIV-1 inhibitors targeting the entrance channel of HIV-1 non-nucleoside reverse transcriptase inhibitor binding pocket.Eur J Med Chem. 2019 Jul 15;174:277-291. doi: 10.1016/j.ejmech.2019.04.054. Epub 2019 Apr 24. Eur J Med Chem. 2019. PMID: 31051402 Review.
Cited by
-
Viral inhibitors derived from macroalgae, microalgae, and cyanobacteria: A review of antiviral potential throughout pathogenesis.Algal Res. 2021 Jul;57:102331. doi: 10.1016/j.algal.2021.102331. Epub 2021 May 18. Algal Res. 2021. PMID: 34026476 Free PMC article.
-
Anti-HIV drug development through computational methods.AAPS J. 2014 Jul;16(4):674-80. doi: 10.1208/s12248-014-9604-9. Epub 2014 Apr 24. AAPS J. 2014. PMID: 24760437 Free PMC article. Review.
-
Seaweeds and Corals from the Brazilian Coast: Review on Biotechnological Potential and Environmental Aspects.Molecules. 2023 May 23;28(11):4285. doi: 10.3390/molecules28114285. Molecules. 2023. PMID: 37298760 Free PMC article. Review.
-
Marine-Derived Diterpenes from 2019 to 2024: Structures, Biological Activities, Synthesis and Potential Applications.Mar Drugs. 2025 Feb 7;23(2):72. doi: 10.3390/md23020072. Mar Drugs. 2025. PMID: 39997196 Free PMC article. Review.
-
Novel Antiretroviral Structures from Marine Organisms.Molecules. 2019 Sep 26;24(19):3486. doi: 10.3390/molecules24193486. Molecules. 2019. PMID: 31561445 Free PMC article. Review.
References
-
- WHO Progress Report 2011: Global HIV/AIDS Response. [(accessed on 14 August 2012)]. Available online: http://www.who.int/hiv/pub/progress_report2011/en/index.html.
-
- Kohlstaedt L.A., Wang J., Friedman J.M., Rice P.A., Steitz T.A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

