A novel cytotoxic sequence contributes to influenza A viral protein PB1-F2 pathogenicity and predisposition to secondary bacterial infection
- PMID: 24173220
- PMCID: PMC3911746
- DOI: 10.1128/JVI.01373-13
A novel cytotoxic sequence contributes to influenza A viral protein PB1-F2 pathogenicity and predisposition to secondary bacterial infection
Abstract
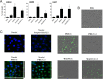
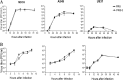
Enhancement of cell death is a distinguishing feature of H1N1 influenza virus A/Puerto Rico/8/34 protein PB1-F2. Comparing the sequences (amino acids [aa] 61 to 87 using PB1-F2 amino acid numbering) of the PB1-F2-derived C-terminal peptides from influenza A viruses inducing high or low levels of cell death, we identified a unique I68, L69, and V70 motif in A/Puerto Rico/8/34 PB1-F2 responsible for promotion of the peptide's cytotoxicity and permeabilization of the mitochondrial membrane. When administered to mice, a 27-mer PB1-F2-derived C-terminal peptide with this amino acid motif caused significantly greater weight loss and pulmonary inflammation than the peptide without it (due to I68T, L69Q, and V70G mutations). Similar to the wild-type peptide, A/Puerto Rico/8/34 elicited significantly higher levels of macrophages, neutrophils, and cytokines in the bronchoalveolar lavage fluid of mice than its mutant counterpart 7 days after infection. Additionally, infection of mice with A/Puerto Rico/8/34 significantly enhanced the levels of morphologically transformed epithelial and immune mononuclear cells recruited in the airways compared with the mutant virus. In the mouse bacterial superinfection model, both peptide and virus with the I68, L69, and V70 sequence accelerated development of pneumococcal pneumonia, as reflected by increased levels of viral and bacterial lung titers and by greater mortality. Here we provide evidence suggesting that the newly identified cytotoxic sequence I68, L69, and V70 of A/Puerto Rico/8/34 PB1-F2 contributes to the pathogenesis of both primary viral and secondary bacterial infections.
Figures






References
-
- Wright PF, Neumann G, Kawaoka Y. 2007. Orthomyxoviruses, p 1691–1740 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

