The evolution of X chromosome inactivation in mammals: the demise of Ohno's hypothesis?
- PMID: 24173285
- PMCID: PMC11113734
- DOI: 10.1007/s00018-013-1499-6
The evolution of X chromosome inactivation in mammals: the demise of Ohno's hypothesis?
Abstract
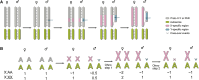
Ohno's hypothesis states that dosage compensation in mammals evolved in two steps: a twofold hyperactivation of the X chromosome in both sexes to compensate for gene losses on the Y chromosome, and silencing of one X (X-chromosome inactivation, XCI) in females to restore optimal dosage. Recent tests of this hypothesis have returned contradictory results. In this review, we explain this ongoing controversy and argue that a novel view on dosage compensation evolution in mammals is starting to emerge. Ohno's hypothesis may be true for a few, dosage-sensitive genes only. If so few genes are compensated, then why has XCI evolved as a chromosome-wide mechanism? This and several other questions raised by the new data in mammals are discussed, and future research directions are proposed.
Figures


References
-
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Marshall Graves JA. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18(6):965–973. doi: 10.1101/gr.7101908. - DOI - PMC - PubMed
-
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

