Improving the representation of peptide-like inhibitor and antibiotic molecules in the Protein Data Bank
- PMID: 24173824
- PMCID: PMC3992913
- DOI: 10.1002/bip.22434
Improving the representation of peptide-like inhibitor and antibiotic molecules in the Protein Data Bank
Abstract
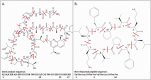
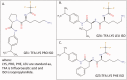
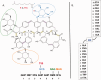
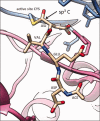
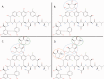
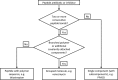
With the accumulation of a large number and variety of molecules in the Protein Data Bank (PDB) comes the need on occasion to review and improve their representation. The Worldwide PDB (wwPDB) partners have periodically updated various aspects of structural data representation to improve the integrity and consistency of the archive. The remediation effort described here was focused on improving the representation of peptide-like inhibitor and antibiotic molecules so that they can be easily identified and analyzed. Peptide-like inhibitors or antibiotics were identified in over 1000 PDB entries, systematically reviewed and represented either as peptides with polymer sequence or as single components. For the majority of the single-component molecules, their peptide-like composition was captured in a new representation, called the subcomponent sequence. A novel concept called "group" was developed for representing complex peptide-like antibiotics and inhibitors that are composed of multiple polymer and nonpolymer components. In addition, a reference dictionary was developed with detailed information about these peptide-like molecules to aid in their annotation, identification and analysis. Based on the experience gained in this remediation, guidelines, procedures, and tools were developed to annotate new depositions containing peptide-like inhibitors and antibiotics accurately and consistently.
Keywords: Protein Data Bank; peptide-like antibiotic; peptide-like inhibitor.
© 2013 The Authors Biopolymers Published by Wiley Periodicals, Inc.
Figures
References
-
- Berman HM, Henrick K, Nakamura H. Nat Struct Biol. 2003;10:980. - PubMed
-
- Henrick K, Feng Z, Bluhm W, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, Lawson CL, Markley JL, Nakamura H, Newman R, Shimizu Y, Swaminathan J, Velankar S, Ory J, Ulrich EL, Vranken W, Westbrook J, Yamashita R, Yang H, Young J, Yousufuddin M, Berman H. Nucleic Acids Res. 2008;36:D426–D433. - PMC - PubMed
-
- Bond CS, Shaw MP, Alphey MS, Hunter WN. Acta Crystallogr Biol Crystallogr. 2001;57:755–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 088944/Wellcome Trust/United Kingdom
- BB/K016970/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I02576X/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- 1R01 GM079429-01A1/GM/NIGMS NIH HHS/United States
- BB/J007471/1/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

