Systems biology of the vervet monkey
- PMID: 24174437
- PMCID: PMC3814400
- DOI: 10.1093/ilar/ilt049
Systems biology of the vervet monkey
Abstract
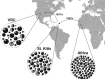
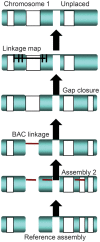
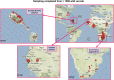
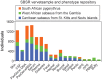
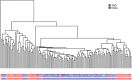
Nonhuman primates (NHP) provide crucial biomedical model systems intermediate between rodents and humans. The vervet monkey (also called the African green monkey) is a widely used NHP model that has unique value for genetic and genomic investigations of traits relevant to human diseases. This article describes the phylogeny and population history of the vervet monkey and summarizes the use of both captive and wild vervet monkeys in biomedical research. It also discusses the effort of an international collaboration to develop the vervet monkey as the most comprehensively phenotypically and genomically characterized NHP, a process that will enable the scientific community to employ this model for systems biology investigations.
Keywords: African green monkey; genetics; genomics; phenomics; simian immunodeficiency virus [SIV]; systems biology; transcriptomics; vervet.
Figures

References
-
- Allen GM. Mammals of the West Indies. Bulletin of the Museum of Comparative Zoology. 1911;54:175–263.
-
- Altmann J, Alberts SC. Growth rates in a wild primate population: Ecological influences and maternal effects. Behav Ecol Sociobiol. 2005;57:490–501.
-
- Bacopoulos NG, Redmond DE, Baulu J, Roth RH. Chronic haloperidol or fluphenazine: Effects on dopamine metabolism in brain, cerebrospinal fluid and plasma of Cercopithecus aethiops (vervet monkey) J Pharmacol Exp Ther 212. 1980:1–5. - PubMed
-
- Bailey JN, Breidenthal SE, Jorgensen MJ, McCracken JT, Fairbanks LA. The association of DRD4 and novelty seeking is found in a nonhuman primate model. Psychiatr Genet. 2007;17:23–27. - PubMed
Publication types
MeSH terms
Grants and funding
- P40RR019963/RR/NCRR NIH HHS/United States
- P40 RR019963/RR/NCRR NIH HHS/United States
- RL1 MH083270/MH/NIMH NIH HHS/United States
- UL1 DE019580/DE/NIDCR NIH HHS/United States
- P30 NS062691/NS/NINDS NIH HHS/United States
- PL1 NS062410/NS/NINDS NIH HHS/United States
- RL1MH083270/MH/NIMH NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- PL1NS062410/NS/NINDS NIH HHS/United States
- UL1DEO19580/PHS HHS/United States
- P30NS062691/NS/NINDS NIH HHS/United States
- R01RR016300/RR/NCRR NIH HHS/United States
- R01 RR016300/RR/NCRR NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

