PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors
- PMID: 24174544
- PMCID: PMC3965000
- DOI: 10.1093/nar/gkt1016
PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors
Abstract
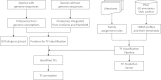
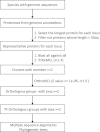
With the aim to provide a resource for functional and evolutionary study of plant transcription factors (TFs), we updated the plant TF database PlantTFDB to version 3.0 (http://planttfdb.cbi.pku.edu.cn). After refining the TF classification pipeline, we systematically identified 129 288 TFs from 83 species, of which 67 species have genome sequences, covering main lineages of green plants. Besides the abundant annotation provided in the previous version, we generated more annotations for identified TFs, including expression, regulation, interaction, conserved elements, phenotype information, expert-curated descriptions derived from UniProt, TAIR and NCBI GeneRIF, as well as references to provide clues for functional studies of TFs. To help identify evolutionary relationship among identified TFs, we assigned 69 450 TFs into 3924 orthologous groups, and constructed 9217 phylogenetic trees for TFs within the same families or same orthologous groups, respectively. In addition, we set up a TF prediction server in this version for users to identify TFs from their own sequences.
Figures

References
-
- Riechmann J, Heard J, Martin G, Reuber L, Keddie J, Adam L, Pineda O, Ratcliffe O, Samaha R, Creelman R. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. - PubMed
-
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics. 2010;26:290–291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

