CXC chemokine receptor 7 (CXCR7) affects the migration of GnRH neurons by regulating CXCL12 availability
- PMID: 24174685
- PMCID: PMC3812513
- DOI: 10.1523/JNEUROSCI.0857-13.2013
CXC chemokine receptor 7 (CXCR7) affects the migration of GnRH neurons by regulating CXCL12 availability
Abstract
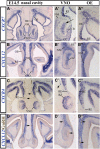
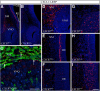
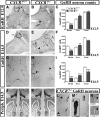
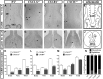
Gonadotropin-releasing hormone (GnRH) neurons are neuroendocrine cells, located in the hypothalamus, that play an essential role in mammalian reproduction. These neurons originate in the nasal placode and migrate during embryonic development, in association with olfactory/vomeronasal nerves, first in the nose, then through the cribriform plate to enter the forebrain, before settling in the hypothalamus. One of the molecules required for their early migration in the nose is the chemokine CXCL12, which is expressed in the embryonic nasal mesenchyme in an increasing ventral to dorsal gradient, presumably guiding GnRH neurons toward the forebrain. Mice lacking CXCR4, the receptor for CXCL12, exhibit defective GnRH cell movement and a significant reduction in their number, suggesting that CXCL12/CXCR4 signaling is important in the migration and survival of these neurons. Here, we investigated the role of the more recently identified second CXCL12 receptor, CXCR7, in GnRH neuron development. We demonstrate that CXCR7 is expressed along the migratory path of GnRH neurons in the nasal cavity and, although not expressed by GnRH neurons, it affects their migration as indicated by the ectopic accumulation of these cells in the nasal compartment in CXCR7(-/-) mice. Absence of CXCR7 caused abnormal accumulation of CXCL12-RFP at CXCR4-positive sites in the nasal area of CXCL12-RFP-transgenic mice and excessive CXCL12-dependent intracellular clustering of CXCR4 in GnRH neurons, suggesting internalization. These findings imply that CXCR7 regulates CXCL12 availability by acting as a scavenger along the migratory path of GnRH neurons and, thus, influences the migration of these cells in a noncell-autonomous manner.
Figures


Similar articles
-
Stromal cell-derived factor-1 (chemokine C-X-C motif ligand 12) and chemokine C-X-C motif receptor 4 are required for migration of gonadotropin-releasing hormone neurons to the forebrain.J Neurosci. 2006 Jun 21;26(25):6834-40. doi: 10.1523/JNEUROSCI.1728-06.2006. J Neurosci. 2006. PMID: 16793890 Free PMC article.
-
CXCR4 and CXCR7 have distinct functions in regulating interneuron migration.Neuron. 2011 Jan 13;69(1):61-76. doi: 10.1016/j.neuron.2010.12.005. Neuron. 2011. PMID: 21220099 Free PMC article.
-
CXC chemokine receptor 7 (CXCR7) regulates CXCR4 protein expression and capillary tuft development in mouse kidney.PLoS One. 2012;7(8):e42814. doi: 10.1371/journal.pone.0042814. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22880115 Free PMC article.
-
Gonadotropin-releasing hormone neuronal migration.Semin Reprod Med. 2007 Sep;25(5):305-12. doi: 10.1055/s-2007-984736. Semin Reprod Med. 2007. PMID: 17710726 Review.
-
The CXCL12-CXCR4/CXCR7 axis as a mechanism of immune resistance in gastrointestinal malignancies.Semin Cancer Biol. 2020 Oct;65:176-188. doi: 10.1016/j.semcancer.2019.12.007. Epub 2019 Dec 23. Semin Cancer Biol. 2020. PMID: 31874281 Review.
Cited by
-
The microcephaly gene Donson is essential for progenitors of cortical glutamatergic and GABAergic neurons.PLoS Genet. 2021 Mar 19;17(3):e1009441. doi: 10.1371/journal.pgen.1009441. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33739968 Free PMC article.
-
Glia Cells Control Olfactory Neurogenesis by Fine-Tuning CXCL12.Cells. 2023 Aug 28;12(17):2164. doi: 10.3390/cells12172164. Cells. 2023. PMID: 37681896 Free PMC article.
-
GnRH-(1-5) Inhibits TGF-β Signaling to Regulate the Migration of Immortalized Gonadotropin-Releasing Hormone Neurons.Front Endocrinol (Lausanne). 2018 Feb 20;9:45. doi: 10.3389/fendo.2018.00045. eCollection 2018. Front Endocrinol (Lausanne). 2018. PMID: 29515521 Free PMC article.
-
ShRNA knock-down of CXCR7 inhibits tumour invasion and metastasis in hepatocellular carcinoma after transcatheter arterial chemoembolization.J Cell Mol Med. 2017 Sep;21(9):1989-1999. doi: 10.1111/jcmm.13119. Epub 2017 Apr 21. J Cell Mol Med. 2017. PMID: 28429395 Free PMC article.
-
Pattern of CXCR7 Gene Expression in Mouse Brain Under Normal and Inflammatory Conditions.J Neuroimmune Pharmacol. 2016 Mar;11(1):26-35. doi: 10.1007/s11481-015-9616-y. Epub 2015 May 22. J Neuroimmune Pharmacol. 2016. PMID: 25997895 Free PMC article.
References
-
- Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ, Wei K, McMaster BE, Wright K, Howard MC, Schall TJ. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
