Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3
- PMID: 24176123
- PMCID: PMC3983652
- DOI: 10.1186/gb-2013-14-10-r121
Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3
Erratum in
-
Erratum to: Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3.Genome Biol. 2016 Feb 4;17:21. doi: 10.1186/s13059-016-0886-3. Genome Biol. 2016. PMID: 26846280 Free PMC article. No abstract available.
Abstract
Background: Nucleosomes are present throughout the genome and must be dynamically regulated to accommodate binding of transcription factors and RNA polymerase machineries by various mechanisms. Despite the development of protocols and techniques that have enabled us to map nucleosome occupancy genome-wide, the dynamic properties of nucleosomes remain poorly understood, particularly in mammalian cells. The histone variant H3.3 is incorporated into chromatin independently of DNA replication and requires displacement of existing nucleosomes for its deposition. Here, we measure H3.3 turnover at high resolution in the mammalian genome in order to present a genome-wide characterization of replication-independent H3.3-nucleosome dynamics.
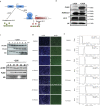
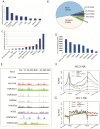
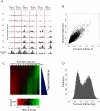
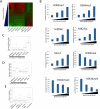
Results: We developed a system to study the DNA replication-independent turnover of nucleosomes containing the histone variant H3.3 in mammalian cells. By measuring the genome-wide incorporation of H3.3 at different time points following epitope-tagged H3.3 expression, we find three categories of H3.3-nucleosome turnover in vivo: rapid turnover, intermediate turnover and, specifically at telomeres, slow turnover. Our data indicate that H3.3-containing nucleosomes at enhancers and promoters undergo rapid turnover that is associated with active histone modification marks including H3K4me1, H3K4me3, H3K9ac, H3K27ac and the histone variant H2A.Z. The rate of turnover is negatively correlated with H3K27me3 at regulatory regions and with H3K36me3 at gene bodies.
Conclusions: We have established a reliable approach to measure turnover rates of H3.3-containing nucleosomes on a genome-wide level in mammalian cells. Our results suggest that distinct mechanisms control the dynamics of H3.3 incorporation at functionally different genomic regions.
Figures


References
-
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;14:180–192. doi: 10.1016/j.stem.2012.11.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

