Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome
- PMID: 24176135
- PMCID: PMC4053723
- DOI: 10.1186/gb-2013-14-11-r122
Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome
Abstract
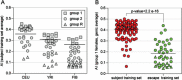
Background: X-chromosome inactivation (XCI) results in the silencing of most genes on one X chromosome, yielding mono-allelic expression in individual cells. However, random XCI results in expression of both alleles in most females. Allelic imbalances have been used genome-wide to detect mono-allelically expressed genes. Analysis of X-linked allelic imbalance in females with skewed XCI offers the opportunity to identify genes that escape XCI with bi-allelic expression in contrast to those with mono-allelic expression and which are therefore subject to XCI.
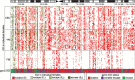
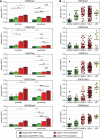
Results: We determine XCI status for 409 genes, all of which have at least five informative females in our dataset. The majority of genes are subject to XCI and genes that escape from XCI show a continuum of expression from the inactive X. Inactive X expression corresponds to differences in the level of histone modification detected by allelic imbalance after chromatin immunoprecipitation. Differences in XCI between populations and between cell lines derived from different tissues are observed.
Conclusions: We demonstrate that allelic imbalance can be used to determine an inactivation status for X-linked genes, even without completely non-random XCI. There is a range of expression from the inactive X. Genes escaping XCI, including those that do so in only a subset of females, cluster together, demonstrating that XCI and location on the X chromosome are related. In addition to revealing mechanisms involved in cis-gene regulation, determining which genes escape XCI can expand our understanding of the contributions of X-linked genes to sexual dimorphism.
Figures


References
-
- Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagné V, Dias J, Hoberman R, Montpetit A, Joly MM, Harvey EJ, Sinnett D, Beaulieu P, Hamon R, Graziani A, Dewar K, Harmsen E, Majewski J, Göring HH, Naumova AK, Blanchette M, Gunderson KL, Pastinen T. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;14:1216–1222. doi: 10.1038/ng.473. - DOI - PubMed
-
- Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010;14:533–538. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

