Loss of iron triggers PINK1/Parkin-independent mitophagy
- PMID: 24176932
- PMCID: PMC3981094
- DOI: 10.1038/embor.2013.168
Loss of iron triggers PINK1/Parkin-independent mitophagy
Abstract
In this study, we develop a simple assay to identify mitophagy inducers on the basis of the use of fluorescently tagged mitochondria that undergo a colour change on lysosomal delivery. Using this assay, we identify iron chelators as a family of compounds that generate a strong mitophagy response. Iron chelation-induced mitophagy requires that cells undergo glycolysis, but does not require PINK1 stabilization or Parkin activation, and occurs in primary human fibroblasts as well as those isolated from a Parkinson's patient with Parkin mutations. Thus, we have identified and characterized a mitophagy pathway, the induction of which could prove beneficial as a potential therapy for several neurodegenerative diseases in which mitochondrial clearance is advantageous.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
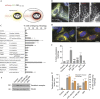
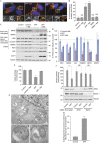
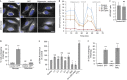
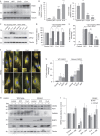
Comment in
-
The many faces of mitophagy.EMBO Rep. 2014 Jan;15(1):5-6. doi: 10.1002/embr.201338224. EMBO Rep. 2014. PMID: 24398127 Free PMC article.
References
-
- Rubinsztein DC, Marino G, Kroemer G (2011) Autophagy and aging. Cell 146: 682–695 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

