Origin and differential selection of allelic variation at TAS2R16 associated with salicin bitter taste sensitivity in Africa
- PMID: 24177185
- PMCID: PMC3907056
- DOI: 10.1093/molbev/mst211
Origin and differential selection of allelic variation at TAS2R16 associated with salicin bitter taste sensitivity in Africa
Abstract
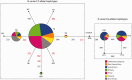
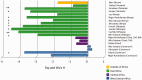
Bitter taste perception influences human nutrition and health, and the genetic variation underlying this trait may play a role in disease susceptibility. To better understand the genetic architecture and patterns of phenotypic variability of bitter taste perception, we sequenced a 996 bp region, encompassing the coding exon of TAS2R16, a bitter taste receptor gene, in 595 individuals from 74 African populations and in 94 non-Africans from 11 populations. We also performed genotype-phenotype association analyses of threshold levels of sensitivity to salicin, a bitter anti-inflammatory compound, in 296 individuals from Central and East Africa. In addition, we characterized TAS2R16 mutants in vitro to investigate the effects of polymorphic loci identified at this locus on receptor function. Here, we report striking signatures of positive selection, including significant Fay and Wu's H statistics predominantly in East Africa, indicating strong local adaptation and greater genetic structure among African populations than expected under neutrality. Furthermore, we observed a "star-like" phylogeny for haplotypes with the derived allele at polymorphic site 516 associated with increased bitter taste perception that is consistent with a model of selection for "high-sensitivity" variation. In contrast, haplotypes carrying the "low-sensitivity" ancestral allele at site 516 showed evidence of strong purifying selection. We also demonstrated, for the first time, the functional effect of nonsynonymous variation at site 516 on salicin phenotypic variance in vivo in diverse Africans and showed that most other nonsynonymous substitutions have weak or no effect on cell surface expression in vitro, suggesting that one main polymorphism at TAS2R16 influences salicin recognition. Additionally, we detected geographic differences in levels of bitter taste perception in Africa not previously reported and infer an East African origin for high salicin sensitivity in human populations.
Keywords: African genetic diversity; genotype–phenotype association; salicin taste perception; selection on standing variation.
Figures



References
-
- Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. - PubMed
-
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. - PubMed
-
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. - PubMed
-
- Barrett RD, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

