Amino acid region 1000-1008 of factor V is a dynamic regulator for the emergence of procoagulant activity
- PMID: 24178294
- PMCID: PMC3873559
- DOI: 10.1074/jbc.M113.462374
Amino acid region 1000-1008 of factor V is a dynamic regulator for the emergence of procoagulant activity
Abstract
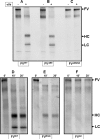
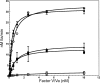
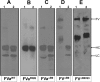
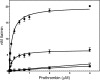
Single chain factor V (fV) circulates as an Mr 330,000 quiescent pro-cofactor. Removal of the B domain and generation of factor Va (fVa) are vital for procoagulant activity. We investigated the role of the basic amino acid region 1000-1008 within the B domain of fV by constructing a recombinant mutant fV molecule with all activation cleavage sites (Arg(709)/Arg(1018)/Arg(1545)) mutated to glutamine (fV(Q3)), a mutant fV molecule with region 1000-1008 deleted (fV(ΔB9)), and a mutant fV molecule containing the same deletion with activation cleavage sites changed to glutamine (fV(ΔB9/Q3)). The recombinant molecules along with wild type fV (fV(WT)) were transiently expressed in COS-7L cells, purified, and assessed for their ability to bind factor Xa (fXa) prior to and following incubation with thrombin. The data showed that fV(Q3) was severely impaired in its interaction with fXa before and after incubation with thrombin. In contrast, KD(app) values for fV(ΔB9) (0.9 nM), fVa(ΔB9) (0.4 nM), and fV(ΔB9/Q3) (0.7 nM) were similar to the affinity of fVa(WT) for fXa (0.3 nM). Two-stage clotting assays revealed that although fV(Q3) was deficient in its clotting activity, fV(ΔB9/Q3) had clotting activity comparable with fVa(WT). The kcat value of prothrombinase assembled with fV(ΔB9/Q3) was minimally affected, whereas the Km value of the reaction was increased 57-fold compared with the Km value obtained with prothrombinase assembled with fVa(WT). These findings strongly suggest that amino acid region 1000-1008 of fV is a regulatory sequence protecting the organisms from spontaneous binding to fXa and unnecessary prothrombinase complex formation, which in turn results in catastrophic physiological consequences.
Keywords: Coagulation Factors; Factor V; Factor Va; Factor Xa; Phospholipid Vesicle; Protein Chemistry; Prothrombin; Prothrombinase; Thrombin.
Figures




References
-
- Mann K. G., Kalafatis M. (2003) Factor V: a combination of Dr. Jekyll and Mr. Hyde. Blood 101, 20–30 - PubMed
-
- Kalafatis M., Egan J. O., van 't Veer C., Cawthern K. M., Mann K. G. (1997) The regulation of clotting factors. Crit. Rev. Eukaryot. Gene Expr. 7, 241–280 - PubMed
-
- Mann K. G., Nesheim M. E., Tracy P. B. (1981) Molecular weight of undegraded plasma factor V. Biochemistry 20, 28–33 - PubMed
-
- Esmon C. T. (1979) The subunit structure of thrombin-activated factor V. Isolation of activated factor V, separation of subunits, and reconstitution of biological activity. J. Biol. Chem. 254, 964–973 - PubMed
-
- Nesheim M. E., Foster W. B., Hewick R., Mann K. G. (1984) Characterization of factor V activation intermediates. J. Biol. Chem. 259, 3187–3196 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

