Histone H4 promotes prothrombin autoactivation
- PMID: 24178300
- PMCID: PMC3861626
- DOI: 10.1074/jbc.M113.509786
Histone H4 promotes prothrombin autoactivation
Abstract
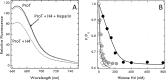
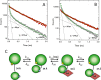
Recent studies have documented the ability of prothrombin to spontaneously convert to the mature protease thrombin when Arg-320 becomes exposed to solvent for proteolytic attack upon mutation of residues in the activation domain. Whether prothrombin autoactivation occurs in the wild-type under conditions relevant to physiology remains unknown. Here, we report that binding of histone H4 to prothrombin under physiological conditions generates thrombin by autoactivation. The effect is abrogated by mutation of the catalytic Ser-525 and requires the presence of the Gla domain. Fluorescence titrations document direct binding of histone H4 to prothrombin with an affinity in the low nm range. Stopped flow data and luminescence resonance energy transfer measurements indicate that the binding mechanism obeys conformational selection. Among the two conformations of prothrombin, collapsed and fully extended, histone H4 binds selectively to the collapsed form and induces a transition toward a new conformation where the distance between Ser-101 in kringle-1 and Ser-210 in kringle-2 increases by 13 Å. These findings confirm the molecular plasticity of prothrombin emerged from recent structural studies and suggest that different conformations of the inter-kringle linker domain determine the functional behavior of prothrombin. The results also broaden our mechanistic understanding of the prothrombotic phenotype observed during cellular damage due to the release of histones in the blood stream. Prothrombin autoactivation induced by histone H4 emerges as a mechanism of pathophysiological relevance through which thrombin is generated independently of activation of the coagulation cascade.
Keywords: Blood Coagulation Factors; Enzyme Mechanisms; Protein Conformation; Prothrombin; Thrombin.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

