Path2Models: large-scale generation of computational models from biochemical pathway maps
- PMID: 24180668
- PMCID: PMC4228421
- DOI: 10.1186/1752-0509-7-116
Path2Models: large-scale generation of computational models from biochemical pathway maps
Abstract
Background: Systems biology projects and omics technologies have led to a growing number of biochemical pathway models and reconstructions. However, the majority of these models are still created de novo, based on literature mining and the manual processing of pathway data.
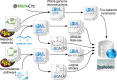
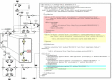
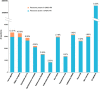
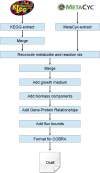
Results: To increase the efficiency of model creation, the Path2Models project has automatically generated mathematical models from pathway representations using a suite of freely available software. Data sources include KEGG, BioCarta, MetaCyc and SABIO-RK. Depending on the source data, three types of models are provided: kinetic, logical and constraint-based. Models from over 2 600 organisms are encoded consistently in SBML, and are made freely available through BioModels Database at http://www.ebi.ac.uk/biomodels-main/path2models. Each model contains the list of participants, their interactions, the relevant mathematical constructs, and initial parameter values. Most models are also available as easy-to-understand graphical SBGN maps.
Conclusions: To date, the project has resulted in more than 140 000 freely available models. Such a resource can tremendously accelerate the development of mathematical models by providing initial starting models for simulation and analysis, which can be subsequently curated and further parameterized.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

