The role of DNA methylation in regulation of the murine Lhx3 gene
- PMID: 24183897
- PMCID: PMC3870101
- DOI: 10.1016/j.gene.2013.10.045
The role of DNA methylation in regulation of the murine Lhx3 gene
Abstract
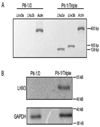
LHX3 is a LIM-homeodomain transcription factor with critical roles in pituitary and nervous system development. Mutations in the LHX3 gene are associated with pediatric diseases featuring severe hormone deficiencies, hearing loss, developmental delay, and other symptoms. The mechanisms that govern LHX3/Lhx3 transcription are poorly understood. In this study, we examined the role of DNA methylation in the expression status of the mouse Lhx3 gene. Pituitary cells that do not normally express Lhx3 (Pit-1/0 cells) were treated with 5-aza-2'-deoxycytidine, a demethylating reagent. This treatment leads to activation of Lhx3 gene expression suggesting that methylation contributes to Lhx3 regulation. Treatment of Pit-1/0 pituitary cells with a combination of a demethylating reagent and a histone deacetylase inhibitor led to rapid activation of Lhx3 expression, suggesting possible crosstalk between DNA methylation and histone modification processes. To assess DNA methylation levels, treated and untreated Pit-1/0 genomic DNAs were subjected to bisulfite conversion and sequencing. Treated Pit-1/0 cells had decreased methylation at specific sites in the Lhx3 locus compared to untreated cells. Chromatin immunoprecipitation assays demonstrated interactions between the MeCp2 methyl binding protein and Lhx3 promoter regions in the Pit-1/0 cell line. Overall, this study demonstrates that DNA methylation patterns of the Lhx3 gene are associated with its expression status.
Keywords: 5-aza-dc; 5′-aza-2′-deoxycytidine; CPHD; Chromatin; CpG; DNA methyltransferase; DNMT; FSHβ; GnRH-R; HDAC; LHX3; LIM-homeodomain protein 3; MBP; NF1; PRL; Promoter; SP1; TSA; TSS; Transcription; alpha glycoprotein subunit; combined pituitary hormone deficiency; cytosine-phosphate bond-guanine; follicle-stimulating hormone; gonadotropin-releasing hormone receptor; histone deacetylase; methyl-CpG binding protein; nuclear factor 1; prolactin; specificity protein 1; transcription start site; trichostatin A; αGSU.
© 2013 Elsevier B.V. All rights reserved.
Figures






Similar articles
-
Characterization of the porcine Lhx3/LIM-3/P-Lim LIM homeodomain transcription factor.Mol Cell Endocrinol. 1999 Jan 25;147(1-2):65-74. doi: 10.1016/s0303-7207(98)00213-5. Mol Cell Endocrinol. 1999. PMID: 10195693
-
LHX3 interacts with inhibitor of histone acetyltransferase complex subunits LANP and TAF-1β to modulate pituitary gene regulation.PLoS One. 2013 Jul 4;8(7):e68898. doi: 10.1371/journal.pone.0068898. Print 2013. PLoS One. 2013. PMID: 23861948 Free PMC article.
-
Regulation of the follicle-stimulating hormone beta gene by the LHX3 LIM-homeodomain transcription factor.Endocrinology. 2004 Nov;145(11):4866-79. doi: 10.1210/en.2004-0598. Epub 2004 Jul 22. Endocrinology. 2004. PMID: 15271874
-
A novel mutation of LHX3 is associated with combined pituitary hormone deficiency including ACTH deficiency, sensorineural hearing loss, and short neck-a case report and review of the literature.Eur J Pediatr. 2011 Aug;170(8):1017-21. doi: 10.1007/s00431-011-1393-x. Epub 2011 Jan 20. Eur J Pediatr. 2011. PMID: 21249393 Review.
-
Roles of the LHX3 and LHX4 LIM-homeodomain factors in pituitary development.Mol Cell Endocrinol. 2007 Feb;265-266:190-5. doi: 10.1016/j.mce.2006.12.019. Epub 2007 Jan 8. Mol Cell Endocrinol. 2007. PMID: 17210222 Free PMC article. Review.
Cited by
-
Differential methylation of genes in individuals exposed to maternal diabetes in utero.Diabetologia. 2017 Apr;60(4):645-655. doi: 10.1007/s00125-016-4203-1. Epub 2017 Jan 26. Diabetologia. 2017. PMID: 28127622 Free PMC article.
-
Epigenetic Modifications in Sensorineural Hearing Loss: Protective Mechanisms and Therapeutic Potential.Curr Med Sci. 2025 Jun;45(3):415-429. doi: 10.1007/s11596-025-00049-9. Epub 2025 May 21. Curr Med Sci. 2025. PMID: 40397300 Review.
-
A Nonsynonymous Substitution of Lhx3 Leads to Changes in Body Size in Dogs and Mice.Genes (Basel). 2024 Jun 4;15(6):739. doi: 10.3390/genes15060739. Genes (Basel). 2024. PMID: 38927675 Free PMC article.
-
Overview of chromatin regulatory processes during surface ectodermal development and homeostasis.Dev Biol. 2024 Nov;515:30-45. doi: 10.1016/j.ydbio.2024.07.001. Epub 2024 Jul 4. Dev Biol. 2024. PMID: 38971398 Review.
References
-
- Auclair G, Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94:2202–2211. - PubMed
-
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. - PubMed
-
- Bhangoo AP, Hunter CS, Savage JJ, Anhalt H, Pavlakis S, Walvoord EC, Ten S, Rhodes SJ. Clinical case seminar: a novel LHX3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocrinol Metab. 2006;91:747–753. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

