Fully automatic segmentation of the mitral leaflets in 3D transesophageal echocardiographic images using multi-atlas joint label fusion and deformable medial modeling
- PMID: 24184435
- PMCID: PMC3897209
- DOI: 10.1016/j.media.2013.10.001
Fully automatic segmentation of the mitral leaflets in 3D transesophageal echocardiographic images using multi-atlas joint label fusion and deformable medial modeling
Abstract
Comprehensive visual and quantitative analysis of in vivo human mitral valve morphology is central to the diagnosis and surgical treatment of mitral valve disease. Real-time 3D transesophageal echocardiography (3D TEE) is a practical, highly informative imaging modality for examining the mitral valve in a clinical setting. To facilitate visual and quantitative 3D TEE image analysis, we describe a fully automated method for segmenting the mitral leaflets in 3D TEE image data. The algorithm integrates complementary probabilistic segmentation and shape modeling techniques (multi-atlas joint label fusion and deformable modeling with continuous medial representation) to automatically generate 3D geometric models of the mitral leaflets from 3D TEE image data. These models are unique in that they establish a shape-based coordinate system on the valves of different subjects and represent the leaflets volumetrically, as structures with locally varying thickness. In this work, expert image analysis is the gold standard for evaluating automatic segmentation. Without any user interaction, we demonstrate that the automatic segmentation method accurately captures patient-specific leaflet geometry at both systole and diastole in 3D TEE data acquired from a mixed population of subjects with normal valve morphology and mitral valve disease.
Keywords: 3D echocardiography; Label fusion; Medial representation; Mitral valve; Multi-atlas segmentation.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures
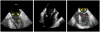
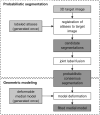







References
-
- Abraham TP, Warner JG, Jr, Kon ND, Lantz PE, Fowle KM, Brooker RF, Ge S, Nomeir AM, Kitzman DW. Feasibility, accuracy, and incremental value of intraoperative three-dimensional transesophageal echocardiography in valve surgery. Am J Cardiol. 1997;80:1577–1582. - PubMed
-
- Ahmed S, Nanda NC, Miller AP, Nekkanti R, Yousif AM, Pacifico AD, Kirklin JK, McGiffin DC. Usefulness of transesophageal three-dimensional echocardiography in the identification of individual segment/scallop prolapse of the mitral valve. Echocardiography. 2003;20:203–209. - PubMed
-
- Artaechevarria X, Munoz-Barrutia A, Ortiz-de-Solorzano C. Combination strategies in multi-atlas image segmentation: application to brain MR data. IEEE Trans Med Imaging. 2009;28:1266–1277. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

