Evolutionary conservation of long non-coding RNAs; sequence, structure, function
- PMID: 24184936
- PMCID: PMC3909678
- DOI: 10.1016/j.bbagen.2013.10.035
Evolutionary conservation of long non-coding RNAs; sequence, structure, function
Abstract
Background: Recent advances in genomewide studies have revealed the abundance of long non-coding RNAs (lncRNAs) in mammalian transcriptomes. The ENCODE Consortium has elucidated the prevalence of human lncRNA genes, which are as numerous as protein-coding genes. Surprisingly, many lncRNAs do not show the same pattern of high interspecies conservation as protein-coding genes. The absence of functional studies and the frequent lack of sequence conservation therefore make functional interpretation of these newly discovered transcripts challenging. Many investigators have suggested the presence and importance of secondary structural elements within lncRNAs, but mammalian lncRNA secondary structure remains poorly understood. It is intriguing to speculate that in this group of genes, RNA secondary structures might be preserved throughout evolution and that this might explain the lack of sequence conservation among many lncRNAs.
Scope of review: Here, we review the extent of interspecies conservation among different lncRNAs, with a focus on a subset of lncRNAs that have been functionally investigated. The function of lncRNAs is widespread and we investigate whether different forms of functionalities may be conserved.
Major conclusions: Lack of conservation does not imbue a lack of function. We highlight several examples of lncRNAs where RNA structure appears to be the main functional unit and evolutionary constraint. We survey existing genomewide studies of mammalian lncRNA conservation and summarize their limitations. We further review specific human lncRNAs which lack evolutionary conservation beyond primates but have proven to be both functional and therapeutically relevant.
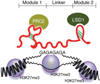
General significance: Pioneering studies highlight a role in lncRNAs for secondary structures, and possibly the presence of functional "modules", which are interspersed with longer and less conserved stretches of nucleotide sequences. Taken together, high-throughput analysis of conservation and functional composition of the still-mysterious lncRNA genes is only now becoming feasible.
Keywords: Antisense RNA; Epigenetic; Evolution; Long non-coding RNA; Polypurines; Secondary structure.
© 2013. Published by Elsevier B.V. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

