Detection of coronaviruses in bats of various species in Italy
- PMID: 24184965
- PMCID: PMC3856409
- DOI: 10.3390/v5112679
Detection of coronaviruses in bats of various species in Italy
Abstract
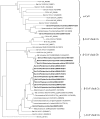
Bats are natural reservoirs for many mammalian coronaviruses, which have received renewed interest after the discovery of the severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) CoV in humans. This study describes the identification and molecular characterization of alphacoronaviruses and betacoronaviruses in bats in Italy, from 2010 to 2012. Sixty-nine faecal samples and 126 carcasses were tested using pan-coronavirus RT-PCR. Coronavirus RNAs were detected in seven faecal samples and nine carcasses. A phylogenetic analysis of RNA-dependent RNA polymerase sequence fragments aided in identifying two alphacoronaviruses from Kuhl's pipistrelle (Pipistrellus kuhlii), three clade 2b betacoronaviruses from lesser horseshoe bats (Rhinolophus hipposideros), and 10 clade 2c betacoronaviruses from Kuhl's pipistrelle, common noctule (Nyctalus noctula), and Savi's pipistrelle (Hypsugo savii). This study fills a substantive gap in the knowledge on bat-CoV ecology in Italy, and extends the current knowledge on clade 2c betacoronaviruses with new sequences obtained from bats that have not been previously described as hosts of these viruses.
Figures
References
-
- De Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A., Holmes K.V., Perlman S., Poon L., Rottier P.J., Talbot P.J., et al. Virus Taxonomy, Classification and Nomenclature of Viruses. Elsevier Academic Press; Philadelphia, PA, USA: 2011. Coronaviridae; pp. 806–828.
-
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wnag M., et al. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

