Activating ESR1 mutations in hormone-resistant metastatic breast cancer
- PMID: 24185510
- PMCID: PMC4009946
- DOI: 10.1038/ng.2823
Activating ESR1 mutations in hormone-resistant metastatic breast cancer
Abstract
Breast cancer is the most prevalent cancer in women, and over two-thirds of cases express estrogen receptor-α (ER-α, encoded by ESR1). Through a prospective clinical sequencing program for advanced cancers, we enrolled 11 patients with ER-positive metastatic breast cancer. Whole-exome and transcriptome analysis showed that six cases harbored mutations of ESR1 affecting its ligand-binding domain (LBD), all of whom had been treated with anti-estrogens and estrogen deprivation therapies. A survey of The Cancer Genome Atlas (TCGA) identified four endometrial cancers with similar mutations of ESR1. The five new LBD-localized ESR1 mutations identified here (encoding p.Leu536Gln, p.Tyr537Ser, p.Tyr537Cys, p.Tyr537Asn and p.Asp538Gly) were shown to result in constitutive activity and continued responsiveness to anti-estrogen therapies in vitro. Taken together, these studies suggest that activating mutations in ESR1 are a key mechanism in acquired endocrine resistance in breast cancer therapy.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


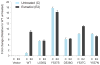
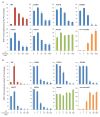
Comment in
-
The search for ESR1 mutations in breast cancer.Nat Genet. 2013 Dec;45(12):1415-6. doi: 10.1038/ng.2831. Nat Genet. 2013. PMID: 24270445 Free PMC article.
-
Drug resistance: making a point.Nat Rev Cancer. 2014 Jan;14(1):6. doi: 10.1038/nrc3649. Epub 2013 Nov 28. Nat Rev Cancer. 2014. PMID: 24285243 No abstract available.
References
-
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. - PubMed
-
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–96. - PubMed
-
- Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

