Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins
- PMID: 24187148
- PMCID: PMC3831976
- DOI: 10.1073/pnas.1318971110
Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins
Abstract
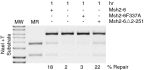
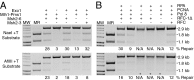
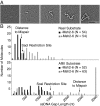
A problem in understanding eukaryotic DNA mismatch repair (MMR) mechanisms is linking insights into MMR mechanisms from genetics and cell-biology studies with those from biochemical studies of MMR proteins and reconstituted MMR reactions. This type of analysis has proven difficult because reconstitution approaches have been most successful for human MMR whereas analysis of MMR in vivo has been most advanced in the yeast Saccharomyces cerevisiae. Here, we describe the reconstitution of MMR reactions using purified S. cerevisiae proteins and mispair-containing DNA substrates. A mixture of MutS homolog 2 (Msh2)-MutS homolog 6, Exonuclease 1, replication protein A, replication factor C-Δ1N, proliferating cell nuclear antigen and DNA polymerase δ was found to repair substrates containing TG, CC, +1 (+T), +2 (+GC), and +4 (+ACGA) mispairs and either a 5' or 3' strand interruption with different efficiencies. The Msh2-MutS homolog 3 mispair recognition protein could substitute for the Msh2-Msh6 mispair recognition protein and showed a different specificity of repair of the different mispairs whereas addition of MutL homolog 1-postmeiotic segregation 1 had no affect on MMR. Repair was catalytic, with as many as 11 substrates repaired per molecule of Exo1. Repair of the substrates containing either a 5' or 3' strand interruption occurred by mispair binding-dependent 5' excision and subsequent resynthesis with excision tracts of up to ~2.9 kb occurring during the repair of the substrate with a 3' strand interruption. The availability of this reconstituted MMR reaction now makes possible detailed biochemical studies of the wealth of mutations identified that affect S. cerevisiae MMR.
Keywords: DNA replication fidelity; cancer; genome instability; mutagenesis; mutator phenotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Reconstitution of Saccharomyces cerevisiae DNA polymerase ε-dependent mismatch repair with purified proteins.Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):3607-3612. doi: 10.1073/pnas.1701753114. Epub 2017 Mar 6. Proc Natl Acad Sci U S A. 2017. PMID: 28265089 Free PMC article.
-
PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair.Mol Cell. 2014 Jul 17;55(2):291-304. doi: 10.1016/j.molcel.2014.04.034. Epub 2014 Jun 26. Mol Cell. 2014. PMID: 24981171 Free PMC article.
-
Activation of Saccharomyces cerevisiae Mlh1-Pms1 Endonuclease in a Reconstituted Mismatch Repair System.J Biol Chem. 2015 Aug 28;290(35):21580-90. doi: 10.1074/jbc.M115.662189. Epub 2015 Jul 13. J Biol Chem. 2015. PMID: 26170454 Free PMC article.
-
Exonuclease 1-dependent and independent mismatch repair.DNA Repair (Amst). 2015 Aug;32:24-32. doi: 10.1016/j.dnarep.2015.04.010. Epub 2015 Apr 30. DNA Repair (Amst). 2015. PMID: 25956862 Free PMC article. Review.
-
Mismatch repair pathway: molecules, functions, and role in colorectal carcinogenesis.Eur J Cancer Prev. 2014 Jul;23(4):246-57. doi: 10.1097/CEJ.0000000000000019. Eur J Cancer Prev. 2014. PMID: 24614649 Review.
Cited by
-
New insights and challenges in mismatch repair: getting over the chromatin hurdle.DNA Repair (Amst). 2014 Jul;19:48-54. doi: 10.1016/j.dnarep.2014.03.027. Epub 2014 Apr 24. DNA Repair (Amst). 2014. PMID: 24767944 Free PMC article. Review.
-
Endonuclease activities of MutLα and its homologs in DNA mismatch repair.DNA Repair (Amst). 2016 Feb;38:42-49. doi: 10.1016/j.dnarep.2015.11.023. Epub 2015 Dec 2. DNA Repair (Amst). 2016. PMID: 26719141 Free PMC article. Review.
-
Schizosaccharomyces pombe MutSα and MutLα Maintain Stability of Tetra-Nucleotide Repeats and Msh3 of Hepta-Nucleotide Repeats.G3 (Bethesda). 2017 May 5;7(5):1463-1473. doi: 10.1534/g3.117.040816. G3 (Bethesda). 2017. PMID: 28341698 Free PMC article.
-
Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex.J Biol Chem. 2014 Mar 28;289(13):9352-64. doi: 10.1074/jbc.M114.552190. Epub 2014 Feb 18. J Biol Chem. 2014. PMID: 24550389 Free PMC article.
-
DNA mismatch repair in the context of chromatin.Cell Biosci. 2020 Feb 3;10:10. doi: 10.1186/s13578-020-0379-7. eCollection 2020. Cell Biosci. 2020. PMID: 32025281 Free PMC article. Review.
References
-
- Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9(1):89–96. - PubMed
-
- Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. - PubMed
-
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

