Oligonucleotide-Based Systems for Input-Controlled and Non-Covalently Regulated Protein-Binding
- PMID: 24187478
- PMCID: PMC3810975
- DOI: 10.1080/10610278.2013.810337
Oligonucleotide-Based Systems for Input-Controlled and Non-Covalently Regulated Protein-Binding
Abstract
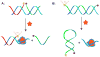
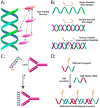
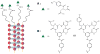
Supramolecular chemists continuously take inspiration from complex biological systems to develop functional molecules involved in molecular recognition and self-assembly. In this regard, "smart" synthetic molecules that emulate allosteric proteins are both exciting and challenging, since many allosteric proteins can be considered as molecular switches that bind to other protein targets in a non-covalent fashion, and importantly, are capable of having their output activity controlled by prior binding to input molecules. This review discusses the foundations and passage toward the development of non-covalently operated oligonucleotide-based systems with protein-binding capacity that can be precisely regulated in an input-controlled manner.
Keywords: input-responsive; multivalency; oligonucleotide; protein binding; structure-switching; supramolecular.
Figures
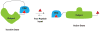

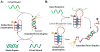
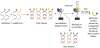
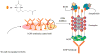
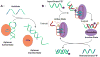




Similar articles
-
Tetrathiafulvalene (TTF)-Annulated Calix[4]pyrroles: Chemically Switchable Systems with Encodable Allosteric Recognition and Logic Gate Functions.Acc Chem Res. 2018 Oct 16;51(10):2400-2410. doi: 10.1021/acs.accounts.8b00308. Epub 2018 Sep 11. Acc Chem Res. 2018. PMID: 30203643
-
Engineering orthogonality in supramolecular polymers: from simple scaffolds to complex materials.Acc Chem Res. 2014 Aug 19;47(8):2405-16. doi: 10.1021/ar500128w. Epub 2014 Jun 6. Acc Chem Res. 2014. PMID: 24905869
-
Molecular Recognition Driven Bioinspired Directional Supramolecular Assembly of Amphiphilic (Macro)molecules and Proteins.Acc Chem Res. 2021 Jun 1;54(11):2670-2682. doi: 10.1021/acs.accounts.1c00195. Epub 2021 May 20. Acc Chem Res. 2021. PMID: 34014638 Review.
-
Supramolecular polymeric materials via cyclodextrin-guest interactions.Acc Chem Res. 2014 Jul 15;47(7):2128-40. doi: 10.1021/ar500109h. Epub 2014 Jun 9. Acc Chem Res. 2014. PMID: 24911321
-
Molecular Recognition in the Colloidal World.Acc Chem Res. 2017 Nov 21;50(11):2756-2766. doi: 10.1021/acs.accounts.7b00370. Epub 2017 Oct 6. Acc Chem Res. 2017. PMID: 28984441 Review.
Cited by
-
Decorating bacteria with self-assembled synthetic receptors.Nat Commun. 2020 Mar 10;11(1):1299. doi: 10.1038/s41467-020-14336-7. Nat Commun. 2020. PMID: 32157077 Free PMC article.
-
Bile Acid Conjugated DNA Chimera that Conditionally Inhibits Carbonic Anhydrase-II in the Presence of MicroRNA-21.Bioconjug Chem. 2015 Aug 19;26(8):1606-12. doi: 10.1021/acs.bioconjchem.5b00231. Epub 2015 Jul 31. Bioconjug Chem. 2015. PMID: 26191606 Free PMC article.
-
Chemically programmable bacterial probes for the recognition of cell surface proteins.Mater Today Bio. 2023 May 23;20:100669. doi: 10.1016/j.mtbio.2023.100669. eCollection 2023 Jun. Mater Today Bio. 2023. PMID: 37334185 Free PMC article.
-
Nucleic Acid Templated Reactions for Chemical Biology.ChemMedChem. 2017 Jun 21;12(12):872-882. doi: 10.1002/cmdc.201700266. ChemMedChem. 2017. PMID: 28480997 Free PMC article. Review.
-
Mimicking the Function of Signaling Proteins: Toward Artificial Signal Transduction Therapy.J Vis Exp. 2016 Sep 29;(115):54396. doi: 10.3791/54396. J Vis Exp. 2016. PMID: 27768030 Free PMC article.
References
-
- Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed. 1998;37(20):2754–2794. - PubMed
-
- Cooper WJ, Waters ML. Curr Opin Chem Biol. 2005;9(6):627–631. - PubMed
-
- Ludden MJW, Reinhoudt DN, Huskens J. Chem Soc Rev. 2006;35(11):1122–1134. - PubMed
-
- Oshovsky GV, Reinhouldt DN, Verboom W. Angew Chem Int Ed. 2007;46:2366–2393. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
