Molecular crowding favors reactivity of a human ribozyme under physiological ionic conditions
- PMID: 24187989
- PMCID: PMC3882164
- DOI: 10.1021/bi400816s
Molecular crowding favors reactivity of a human ribozyme under physiological ionic conditions
Abstract
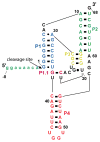
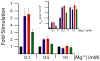
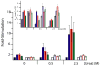
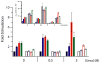
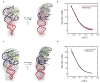
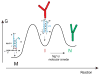
In an effort to relate RNA folding to function under cellular-like conditions, we monitored the self-cleavage reaction of the human hepatitis delta virus-like CPEB3 ribozyme in the background of physiological ionic concentrations and various crowding and cosolute agents. We found that at physiological free Mg(2+) concentrations (∼0.1-0.5 mM), both crowders and cosolutes stimulate the rate of self-cleavage, up to ∼6-fold, but that in 10 mM Mg(2+) (conditions widely used for in vitro ribozyme studies) these same additives have virtually no effect on the self-cleavage rate. We further observe a dependence of the self-cleavage rate on crowder size, wherein the level of rate stimulation is diminished for crowders larger than the size of the unfolded RNA. Monitoring effects of crowding and cosolute agents on rates in biological amounts of urea revealed additive-promoted increases at both low and high Mg(2+) concentrations, with a maximal stimulation of more than 10-fold and a rescue of the rate to its urea-free values. Small-angle X-ray scattering experiments reveal a structural basis for this stimulation in that higher-molecular weight crowding agents favor a more compact form of the ribozyme in 0.5 mM Mg(2+) that is essentially equivalent to the form under standard ribozyme conditions of 10 mM Mg(2+) without a crowder. This finding suggests that at least a portion of the rate enhancement arises from favoring the native RNA tertiary structure. We conclude that cellular-like crowding supports ribozyme reactivity by favoring a compact form of the ribozyme, but only under physiological ionic and cosolute conditions.
Figures

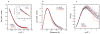
References
-
- Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nat Chem. 2012;4:941. - PubMed
-
- Minton AP. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J Biol Chem. 2001;276:10577–10580. - PubMed
-
- Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. - PubMed
-
- Nakano S, Karimata HT, Kitagawa Y, Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J Am Chem Soc. 2009;131:16881–16888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

