The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase
- PMID: 24189073
- PMCID: PMC3861651
- DOI: 10.1074/jbc.M113.525618
The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase
Abstract
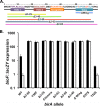
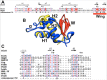
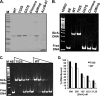
The BirA biotin protein ligase of Escherichia coli belongs to the winged helix-turn-helix (wHTH) family of transcriptional regulators. The N-terminal BirA domain is required for both transcriptional regulation of biotin synthesis and biotin protein ligase activity. We addressed the structural and functional role of the wing of the wHTH motif in both BirA functions. A panel of N-terminal deletion mutant proteins including a discrete deletion of the wing motif were unable to bind DNA. However, all the N-terminal deletion mutants weakly complemented growth of a ΔbirA strain at low biotin concentrations, indicating compromised ligase activity. A wing domain chimera was constructed by replacing the BirA wing with the nearly isosteric wing of the E. coli OmpR transcription factor. Although this chimera BirA was defective in operator binding, it was much more efficient in complementation of a ΔbirA strain than was the wing-less protein. The enzymatic activities of the wing deletion and chimera proteins in the in vitro synthesis of biotinoyl-5'-AMP differed greatly. The wing deletion BirA accumulated an off pathway compound, ADP, whereas the chimera protein did not. Finally, we report that a single residue alteration in the wing bypasses the deleterious effects caused by mutations in the biotin-binding loop of the ligase active site. We believe that the role of the wing in the BirA enzymatic reaction is to orient the active site and thereby protect biotinoyl-5'-AMP from attack by solvent. This is the first evidence that the wing domain of a wHTH protein can play an important role in enzymatic activity.
Keywords: AMP; Bacterial Genetics; Bacterial Metabolism; Enzyme Catalysis; Enzyme Mutation; Microbiology; Promoters; Transcription Factors; Transcription Regulation; Transcription Target Genes.
Figures




References
-
- Beckett D. (2007) Biotin sensing. Universal influence of biotin status on transcription. Annu. Rev. Genet. 41, 443–464 - PubMed
-
- Cronan J. E., Jr. (1988) Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J. Biol. Chem. 263, 10332–10336 - PubMed
-
- Streaker E. D., Gupta A., Beckett D. (2002) The biotin repressor. Thermodynamic coupling of corepressor binding, protein assembly, and sequence-specific DNA binding. Biochemistry 41, 14263–14271 - PubMed
-
- Mukhopadhyay B., Purwantini E., Kreder C. L., Wolfe R. S. (2001) Oxaloacetate synthesis in the methanarchaeon Methanosarcina barkeri. Pyruvate carboxylase genes and a putative Escherichia coli-type bifunctional biotin protein ligase gene (bpl/birA) exhibit a unique organization. J. Bacteriol. 183, 3804–3810 - PMC - PubMed
-
- Streaker E. D., Beckett D. (1998) A map of the biotin repressor-biotin operator interface. Binding of a winged helix-turn-helix protein dimer to a forty base-pair site. J. Mol. Biol. 278, 787–800 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

