Molecular replacement: tricks and treats
- PMID: 24189227
- PMCID: PMC3817689
- DOI: 10.1107/S0907444913015291
Molecular replacement: tricks and treats
Abstract
Molecular replacement is the method of choice for X-ray crystallographic structure determination provided that suitable structural homologues are available in the PDB. Presently, there are ~80,000 structures in the PDB (8074 were deposited in the year 2012 alone), of which ~70% have been solved by molecular replacement. For successful molecular replacement the model must cover at least 50% of the total structure and the Cα r.m.s.d. between the core model and the structure to be solved must be less than 2 Å. Here, an approach originally implemented in the CaspR server (http://www.igs.cnrs-mrs.fr/Caspr2/index.cgi) based on homology modelling to search for a molecular-replacement solution is discussed. How the use of as much information as possible from different sources can improve the model(s) is briefly described. The combination of structural information with distantly related sequences is crucial to optimize the multiple alignment that will define the boundaries of the core domains. PDB clusters (sequences with ≥30% identical residues) can also provide information on the eventual changes in conformation and will help to explore the relative orientations assumed by protein subdomains. Normal-mode analysis can also help in generating series of conformational models in the search for a molecular-replacement solution. Of course, finding a correct solution is only the first step and the accuracy of the identified solution is as important as the data quality to proceed through refinement. Here, some possible reasons for failure are discussed and solutions are proposed using a set of successful examples.
Keywords: molecular replacement.
Figures
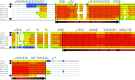

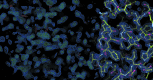
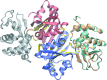
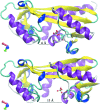
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

