Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens
- PMID: 24189654
- PMCID: PMC3873496
- DOI: 10.1016/j.jmoldx.2013.08.004
Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens
Abstract
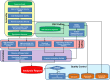
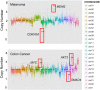
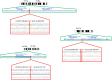
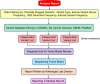
Recent years have seen development and implementation of anticancer therapies targeted to particular gene mutations, but methods to assay clinical cancer specimens in a comprehensive way for the critical mutations remain underdeveloped. We have developed UW-OncoPlex, a clinical molecular diagnostic assay to provide simultaneous deep-sequencing information, based on >500× average coverage, for all classes of mutations in 194 clinically relevant genes. To validate UW-OncoPlex, we tested 98 previously characterized clinical tumor specimens from 10 different cancer types, including 41 formalin-fixed paraffin-embedded tissue samples. Mixing studies indicated reliable mutation detection in samples with ≥ 10% tumor cells. In clinical samples with ≥ 10% tumor cells, UW-OncoPlex correctly identified 129 of 130 known mutations [sensitivity 99.2%, (95% CI, 95.8%-99.9%)], including single nucleotide variants, small insertions and deletions, internal tandem duplications, gene copy number gains and amplifications, gene copy losses, chromosomal gains and losses, and actionable genomic rearrangements, including ALK-EML4, ROS1, PML-RARA, and BCR-ABL. In the same samples, the assay also identified actionable point mutations in genes not previously analyzed and novel gene rearrangements of MLL and GRIK4 in melanoma, and of ASXL1, PIK3R1, and SGCZ in acute myeloid leukemia. To best guide existing and emerging treatment regimens and facilitate integration of genomic testing with patient care, we developed a framework for data analysis, decision support, and reporting clinically actionable results.
Copyright © 2014 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Slamon D.J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. - PubMed
-
- Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., Louis D.N., Christiani D.C., Settleman J., Haber D.A. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., Naoki K., Sasaki H., Fujii Y., Eck M.J., Sellers W.R., Johnson B.E., Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
-
- Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. - PMC - PubMed
-
- Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O’Day S.J., Sosman J.A., Kirkwood J.M., Eggermont A.M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R.J., Flaherty K.T., McArthur G.A. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

