Genome-wide association study of subtype-specific epithelial ovarian cancer risk alleles using pooled DNA
- PMID: 24190013
- PMCID: PMC4063682
- DOI: 10.1007/s00439-013-1383-3
Genome-wide association study of subtype-specific epithelial ovarian cancer risk alleles using pooled DNA
Abstract
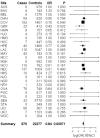
Epithelial ovarian cancer (EOC) is a heterogeneous cancer with both genetic and environmental risk factors. Variants influencing the risk of developing the less-common EOC subtypes have not been fully investigated. We performed a genome-wide association study (GWAS) of EOC according to subtype by pooling genomic DNA from 545 cases and 398 controls of European descent, and testing for allelic associations. We evaluated for replication 188 variants from the GWAS [56 variants for mucinous, 55 for endometrioid and clear cell, 53 for low-malignant potential (LMP) serous, and 24 for invasive serous EOC], selected using pre-defined criteria. Genotypes from 13,188 cases and 23,164 controls of European descent were used to perform unconditional logistic regression under the log-additive genetic model; odds ratios (OR) and 95 % confidence intervals are reported. Nine variants tagging six loci were associated with subtype-specific EOC risk at P < 0.05, and had an OR that agreed in direction of effect with the GWAS results. Several of these variants are in or near genes with a biological rationale for conferring EOC risk, including ZFP36L1 and RAD51B for mucinous EOC (rs17106154, OR = 1.17, P = 0.029, n = 1,483 cases), GRB10 for endometrioid and clear cell EOC (rs2190503, P = 0.014, n = 2,903 cases), and C22orf26/BPIL2 for LMP serous EOC (rs9609538, OR = 0.86, P = 0.0043, n = 892 cases). In analyses that included the 75 GWAS samples, the association between rs9609538 (OR = 0.84, P = 0.0007) and LMP serous EOC risk remained statistically significant at P < 0.0012 adjusted for multiple testing. Replication in additional samples will be important to verify these results for the less-common EOC subtypes.
Figures



References
-
- Albarracin CT, Silva EG, Malpica A, Luthra R, Liu J. The role of hMSH3 and hMSH6 in ovarian endometrioid carcinoma and relationship with microsatellite instability phenotype. Oncol Rep. 2004;12:1217–9. - PubMed
-
- Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, Hillman KM, Mai PL, Lawrenson K, Stutz MD, Lu Y, Karevan R, Woods N, Johnston RL, French JD, Chen X, Weischer M, Nielsen SF, Maranian MJ, Ghoussaini M, Ahmed S, Baynes C, Bolla MK, Wang Q, Dennis J, McGuffog L, Barrowdale D, Lee A, Healey S, Lush M, Tessier DC, Vincent D, Bacot F, Vergote I, Lambrechts S, Despierre E, Risch HA, Gonzalez-Neira A, Rossing MA, Pita G, Doherty JA, Alvarez N, Larson MC, Fridley BL, Schoof N, Chang-Claude J, Cicek MS, Peto J, Kalli KR, Broeks A, Armasu SM, Schmidt MK, Braaf LM, Winterhoff B, Nevanlinna H, Konecny GE, Lambrechts D, Rogmann L, Guenel P, Teoman A, Milne RL, Garcia JJ, Cox A, Shridhar V, Burwinkel B, Marme F, Hein R, Sawyer EJ, Haiman CA, Wang-Gohrke S, Andrulis IL, Moysich KB, Hopper JL, Odunsi K, Lindblom A, Giles GG, Brenner H, Simard J, Lurie G, Fasching PA, Carney ME, Radice P, Wilkens LR, Swerdlow A, Goodman MT, Brauch H, Garcia-Closas M, Hillemanns P, Winqvist R, Durst M, Devilee P, Runnebaum I, Jakubowska A, Lubinski J, Mannermaa A, Butzow R, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–84. 384e1–2. doi: 10.1038/ng.2566ng.2566[pii]. - DOI - PMC - PubMed
-
- Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, Weidhaas J, Paik D, Van Den Berg DJ, Stram DO, Pearce CL, Wu AH, Brewster W, Anton-Culver H, Ziogas A, Narod SA, Levine DA, Kaye SB, Brown R, Paul J, Flanagan J, Sieh W, McGuire V, Whittemore AS, Campbell I, Gore ME, Lissowska J, Yang HP, Medrek K, Gronwald J, Lubinski J, Jakubowska A, Le ND, Cook LS, Kelemen LE, Brook-Wilson A, Massuger LF, Kiemeney LA, Aben KK, van Altena AM, Houlston R, Tomlinson I, Palmieri RT, Moorman PG, Schildkraut J, Iversen ES, Phelan C, Vierkant RA, Cunningham JM, Goode EL, Fridley BL, Kruger-Kjaer S, Blaeker J, Hogdall E, Hogdall C, Gross J, Karlan BY, Ness RB, Edwards RP, Odunsi K, Moyisch KB, Baker JA, Modugno F, Heikkinenen T, Butzow R, Nevanlinna H, Leminen A, Bogdanova N, Antonenkova N, Doerk T, Hillemanns P, Durst M, Runnebaum I, Thompson PJ, Carney ME, Goodman MT, Lurie G, Wang-Gohrke S, Hein R, Chang-Claude J, Rossing MA, Cushing-Haugen KL, Doherty J, Chen C, Rafnar T, Besenbacher S, Sulem P, Stefansson K, Birrer MJ, Terry KL, Hernandez D, Cramer DW, Vergote I, Amant F, Lambrechts D, Despierre E, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nature genetics. 2010;42:880–884. doi: 10.1038/ng.666. - DOI - PMC - PubMed
-
- Brown KM, Macgregor S, Montgomery GW, Craig DW, Zhao ZZ, Iyadurai K, Henders AK, Homer N, Campbell MJ, Stark M, Thomas S, Schmid H, Holland EA, Gillanders EM, Duffy DL, Maskiell JA, Jetann J, Ferguson M, Stephan DA, Cust AE, Whiteman D, Green A, Olsson H, Puig S, Ghiorzo P, Hansson J, Demenais F, Goldstein AM, Gruis NA, Elder DE, Bishop JN, Kefford RF, Giles GG, Armstrong BK, Aitken JF, Hopper JL, Martin NG, Trent JM, Mann GJ, Hayward NK. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nature genetics. 2008;40:838–840. doi: 10.1038/ng.163. - DOI - PMC - PubMed
-
- Cai KQ, Albarracin C, Rosen D, Zhong R, Zheng W, Luthra R, Broaddus R, Liu J. Microsatellite instability and alteration of the expression of hMLH1 and hMSH2 in ovarian clear cell carcinoma. Hum Pathol. 2004;35:552–9. doi:S0046817704000450[pii] - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R03-CA115195/CA/NCI NIH HHS/United States
- P50 CA105009/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- R01-CA092044/CA/NCI NIH HHS/United States
- U01 CA069417/CA/NCI NIH HHS/United States
- R01 CA050385/CA/NCI NIH HHS/United States
- R01-CA067262/CA/NCI NIH HHS/United States
- N01 PC067010/PC/NCI NIH HHS/United States
- P01-CA017054/CA/NCI NIH HHS/United States
- C536/A13086/CRUK_/Cancer Research UK/United Kingdom
- P50 CA159981/CA/NCI NIH HHS/United States
- C490/A6187/CRUK_/Cancer Research UK/United Kingdom
- P30 CA016056/CA/NCI NIH HHS/United States
- R01 CA087538/CA/NCI NIH HHS/United States
- R01 CA136924/CA/NCI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- K22-CA138563/CA/NCI NIH HHS/United States
- R01 CA067262/CA/NCI NIH HHS/United States
- R01 CA106414/CA/NCI NIH HHS/United States
- M01 RR000056/RR/NCRR NIH HHS/United States
- C490/A10119/CRUK_/Cancer Research UK/United Kingdom
- C536/A6689/CRUK_/Cancer Research UK/United Kingdom
- N01-CN55424/CN/NCI NIH HHS/United States
- R01-CA054419/CA/NCI NIH HHS/United States
- R03-CA113148/CA/NCI NIH HHS/United States
- MOP-84340/CAPMC/ CIHR/Canada
- R03 CA113148/CA/NCI NIH HHS/United States
- R01 CA058598/CA/NCI NIH HHS/United States
- MO1-RR000056/RR/NCRR NIH HHS/United States
- R37-CA070867/CA/NCI NIH HHS/United States
- R37 CA070867/CA/NCI NIH HHS/United States
- R01-CA112523/CA/NCI NIH HHS/United States
- R01-CA049449/CA/NCI NIH HHS/United States
- R01-CA080742/CA/NCI NIH HHS/United States
- K22 CA138563/CA/NCI NIH HHS/United States
- 11022/CRUK_/Cancer Research UK/United Kingdom
- N01-PC067010/PC/NCI NIH HHS/United States
- R01-CA106414/CA/NCI NIH HHS/United States
- R01 CA058860/CA/NCI NIH HHS/United States
- R01 CA080742/CA/NCI NIH HHS/United States
- R01-CA126841/CA/NCI NIH HHS/United States
- R01-CA058598/CA/NCI NIH HHS/United States
- R01 CA074850/CA/NCI NIH HHS/United States
- R01-CA122443/CA/NCI NIH HHS/United States
- R01 CA063678/CA/NCI NIH HHS/United States
- K07 CA092044/CA/NCI NIH HHS/United States
- R01-CA087538/CA/NCI NIH HHS/United States
- R01-CA014089/CA/NCI NIH HHS/United States
- K07 CA080668/CA/NCI NIH HHS/United States
- R01-CA074850/CA/NCI NIH HHS/United States
- C490/A10124/CRUK_/Cancer Research UK/United Kingdom
- P30 CA008748/CA/NCI NIH HHS/United States
- R01-CA061132/CA/NCI NIH HHS/United States
- R01-CA064277/CA/NCI NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- R01 CA064277/CA/NCI NIH HHS/United States
- R01-CA050385/CA/NCI NIH HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- MOP-86727/CAPMC/ CIHR/Canada
- P50-CA105009/CA/NCI NIH HHS/United States
- R01 CA083918/CA/NCI NIH HHS/United States
- U01-CA069417/CA/NCI NIH HHS/United States
- R01-CA136924/CA/NCI NIH HHS/United States
- R03 CA115195/CA/NCI NIH HHS/United States
- U01-CA071966/CA/NCI NIH HHS/United States
- R01-CA017054/CA/NCI NIH HHS/United States
- R01 CA054419/CA/NCI NIH HHS/United States
- R01 CA122443/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- R01-CA016056/CA/NCI NIH HHS/United States
- R01 CA076016/CA/NCI NIH HHS/United States
- R01-CA083918/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- K07 CA143047/CA/NCI NIH HHS/United States
- K07-CA143047/CA/NCI NIH HHS/United States
- R01 CA087696/CA/NCI NIH HHS/United States
- R01-CA149429/CA/NCI NIH HHS/United States
- R01 CA149429/CA/NCI NIH HHS/United States
- R01-CA071766/CA/NCI NIH HHS/United States
- P01 CA017054/CA/NCI NIH HHS/United States
- P01-CA087696/CA/NCI NIH HHS/United States
- 13086/CRUK_/Cancer Research UK/United Kingdom
- N01 CN025403/CA/NCI NIH HHS/United States
- 10119/CRUK_/Cancer Research UK/United Kingdom
- R01-CA114343/CA/NCI NIH HHS/United States
- R01 CA049449/CA/NCI NIH HHS/United States
- R01-CA076016/CA/NCI NIH HHS/United States
- R37-CA70867/CA/NCI NIH HHS/United States
- K07-CA095666/CA/NCI NIH HHS/United States
- R01 CA063682/CA/NCI NIH HHS/United States
- K07-CA080668/CA/NCI NIH HHS/United States
- R01-CA061107/CA/NCI NIH HHS/United States
- P50-CA136393/CA/NCI NIH HHS/United States
- K07 CA095666/CA/NCI NIH HHS/United States
- 10118/CRUK_/Cancer Research UK/United Kingdom
- N01-PC035137/PC/NCI NIH HHS/United States
- R01 CA112523/CA/NCI NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
- R01 CA126841/CA/NCI NIH HHS/United States
- P30-CA15083/CA/NCI NIH HHS/United States
- R01 CA114343/CA/NCI NIH HHS/United States
- R01-CA058860/CA/NCI NIH HHS/United States
- R01-CA080978/CA/NCI NIH HHS/United States
- 16561/CRUK_/Cancer Research UK/United Kingdom
- 10124/CRUK_/Cancer Research UK/United Kingdom
- N01 PC035137/PC/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

