A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis
- PMID: 24190431
- PMCID: PMC3832918
- DOI: 10.1084/jem.20131241
A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis
Abstract
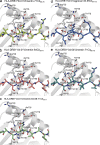
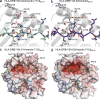
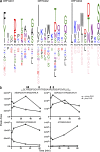
Rheumatoid arthritis (RA) is strongly associated with the human leukocyte antigen (HLA)-DRB1 locus that possesses the shared susceptibility epitope (SE) and the citrullination of self-antigens. We show how citrullinated aggrecan and vimentin epitopes bind to HLA-DRB1*04:01/04. Citrulline was accommodated within the electropositive P4 pocket of HLA-DRB1*04:01/04, whereas the electronegative P4 pocket of the RA-resistant HLA-DRB1*04:02 allomorph interacted with arginine or citrulline-containing epitopes. Peptide elution studies revealed P4 arginine-containing peptides from HLA-DRB1*04:02, but not from HLA-DRB1*04:01/04. Citrullination altered protease susceptibility of vimentin, thereby generating self-epitopes that are presented to T cells in HLA-DRB1*04:01(+) individuals. Using HLA-II tetramers, we observed citrullinated vimentin- and aggrecan-specific CD4(+) T cells in the peripheral blood of HLA-DRB1*04:01(+) RA-affected and healthy individuals. In RA patients, autoreactive T cell numbers correlated with disease activity and were deficient in regulatory T cells relative to healthy individuals. These findings reshape our understanding of the association between citrullination, the HLA-DRB1 locus, and T cell autoreactivity in RA.
Figures



References
-
- Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O., III, Birnbaum N.S., Burmester G.R., Bykerk V.P., Cohen M.D., et al. 2010. Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 62:2569–2581 10.1002/art.27584 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

