Ribosome biogenesis in the yeast Saccharomyces cerevisiae
- PMID: 24190922
- PMCID: PMC3813855
- DOI: 10.1534/genetics.113.153197
Ribosome biogenesis in the yeast Saccharomyces cerevisiae
Abstract
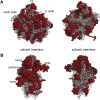
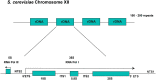
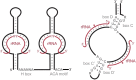
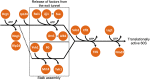
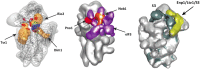
Ribosomes are highly conserved ribonucleoprotein nanomachines that translate information in the genome to create the proteome in all cells. In yeast these complex particles contain four RNAs (>5400 nucleotides) and 79 different proteins. During the past 25 years, studies in yeast have led the way to understanding how these molecules are assembled into ribosomes in vivo. Assembly begins with transcription of ribosomal RNA in the nucleolus, where the RNA then undergoes complex pathways of folding, coupled with nucleotide modification, removal of spacer sequences, and binding to ribosomal proteins. More than 200 assembly factors and 76 small nucleolar RNAs transiently associate with assembling ribosomes, to enable their accurate and efficient construction. Following export of preribosomes from the nucleus to the cytoplasm, they undergo final stages of maturation before entering the pool of functioning ribosomes. Elaborate mechanisms exist to monitor the formation of correct structural and functional neighborhoods within ribosomes and to destroy preribosomes that fail to assemble properly. Studies of yeast ribosome biogenesis provide useful models for ribosomopathies, diseases in humans that result from failure to properly assemble ribosomes.
Keywords: RNA polymerase I; ribosomal RNA; ribosome; ribosome assembly; snoRNAs.
Figures
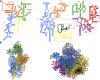


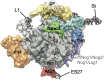
References
-
- Allmang C., Tollervey D., 1998. The role of the 3′ external transcribed spacer in yeast pre- rRNA processing. J. Mol. Biol. 278: 67–78 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

