A role for USP7 in DNA replication
- PMID: 24190967
- PMCID: PMC3911275
- DOI: 10.1128/MCB.00639-13
A role for USP7 in DNA replication
Abstract
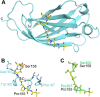
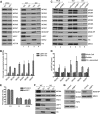
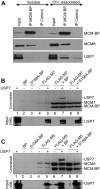
The minichromosome maintenance (MCM) complex, which plays multiple important roles in DNA replication, is loaded onto chromatin following mitosis, remains on chromatin until the completion of DNA synthesis, and then is unloaded by a poorly defined mechanism that involves the MCM binding protein (MCM-BP). Here we show that MCM-BP directly interacts with the ubiquitin-specific protease USP7, that this interaction occurs predominantly on chromatin, and that MCM-BP can tether USP7 to MCM proteins. Detailed biochemical and structure analyses of the USP7-MCM-BP interaction showed that the (155)PSTS(158) MCM-BP sequence mediates critical interactions with the TRAF domain binding pocket of USP7. Analysis of the effects of USP7 knockout on DNA replication revealed that lack of USP7 results in slowed progression through late S phase without globally affecting the fork rate or origin usage. Lack of USP7 also resulted in increased levels of MCM proteins on chromatin, and investigation of the cause of this increase revealed a defect in the dissociation of MCM proteins from chromatin in mid- to late S phase. This role of USP7 mirrors the previously described role for MCM-BP in MCM complex unloading and suggests that USP7 works with MCM-BP to unload MCM complexes from chromatin at the end of S phase.
Figures






Similar articles
-
Identification and characterization of a novel component of the human minichromosome maintenance complex.Mol Cell Biol. 2007 Apr;27(8):3044-55. doi: 10.1128/MCB.02384-06. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296731 Free PMC article.
-
Interaction of human minichromosome maintenance protein-binding protein with minichromosome maintenance 2-7.FEBS J. 2014 Feb;281(4):1057-67. doi: 10.1111/febs.12668. Epub 2014 Jan 15. FEBS J. 2014. PMID: 24299456
-
Interactions of the human MCM-BP protein with MCM complex components and Dbf4.PLoS One. 2012;7(4):e35931. doi: 10.1371/journal.pone.0035931. Epub 2012 Apr 23. PLoS One. 2012. PMID: 22540012 Free PMC article.
-
The MCM complex: its role in DNA replication and implications for cancer therapy.Curr Cancer Drug Targets. 2005 Aug;5(5):365-80. doi: 10.2174/1568009054629654. Curr Cancer Drug Targets. 2005. PMID: 16101384 Review.
-
Molecular Mechanism for Chromatin Regulation During MCM Loading in Mammalian Cells.Adv Exp Med Biol. 2017;1042:61-78. doi: 10.1007/978-981-10-6955-0_3. Adv Exp Med Biol. 2017. PMID: 29357053 Review.
Cited by
-
The termination of UHRF1-dependent PAF15 ubiquitin signaling is regulated by USP7 and ATAD5.Elife. 2023 Feb 3;12:e79013. doi: 10.7554/eLife.79013. Elife. 2023. PMID: 36734974 Free PMC article.
-
USP7: Structure, substrate specificity, and inhibition.DNA Repair (Amst). 2019 Apr;76:30-39. doi: 10.1016/j.dnarep.2019.02.005. Epub 2019 Feb 16. DNA Repair (Amst). 2019. PMID: 30807924 Free PMC article. Review.
-
Structural Characterization of Interaction between Human Ubiquitin-specific Protease 7 and Immediate-Early Protein ICP0 of Herpes Simplex Virus-1.J Biol Chem. 2015 Sep 18;290(38):22907-18. doi: 10.1074/jbc.M115.664805. Epub 2015 Jul 29. J Biol Chem. 2015. PMID: 26224631 Free PMC article.
-
Epstein-Barr Virus BGLF2 commandeers RISC to interfere with cellular miRNA function.PLoS Pathog. 2022 Jan 10;18(1):e1010235. doi: 10.1371/journal.ppat.1010235. eCollection 2022 Jan. PLoS Pathog. 2022. PMID: 35007297 Free PMC article.
-
USP7 regulates the ERK1/2 signaling pathway through deubiquitinating Raf-1 in lung adenocarcinoma.Cell Death Dis. 2022 Aug 10;13(8):698. doi: 10.1038/s41419-022-05136-6. Cell Death Dis. 2022. PMID: 35948545 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
