Regulatory R region of the CFTR chloride channel is a dynamic integrator of phospho-dependent intra- and intermolecular interactions
- PMID: 24191035
- PMCID: PMC3839774
- DOI: 10.1073/pnas.1315104110
Regulatory R region of the CFTR chloride channel is a dynamic integrator of phospho-dependent intra- and intermolecular interactions
Abstract
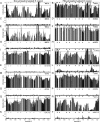
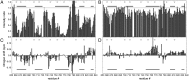
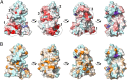
Intrinsically disordered proteins play crucial roles in regulatory processes and often function as protein interaction hubs. Here, we present a detailed characterization of a full-length disordered hub protein region involved in multiple dynamic complexes. We performed NMR, CD, and fluorescence binding studies on the nonphosphorylated and highly PKA-phosphorylated human cystic fibrosis transmembrane conductance regulator (CFTR) regulatory region, a ∼200-residue disordered segment involved in phosphorylation-dependent regulation of channel trafficking and gating. Our data provide evidence for dynamic, phosphorylation-dependent, multisite interactions of various segments of the regulatory region for its intra- and intermolecular partners, including the CFTR nucleotide binding domains 1 and 2, a 42-residue peptide from the C terminus of CFTR, the SLC26A3 sulphate transporter and antisigma factor antagonist (STAS) domain, and 14-3-3β. Because of its large number of binding partners, multivalent binding of individually weak sites facilitates rapid exchange between free and bound states to allow the regulatory region to engage with different partners and generate a graded or rheostat-like response to phosphorylation. Our results enrich the understanding of how disordered binding segments interact with multiple targets. We present structural models consistent with our data that illustrate this dynamic aspect of phospho-regulation of CFTR by the disordered regulatory region.
Keywords: fuzzy complex; protein interaction network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. - PubMed
-
- Tompa P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem Sci. 2012;37(12):509–516. - PubMed
-
- Dosztányi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res. 2006;5(11):2985–2995. - PubMed
-
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

