Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET
- PMID: 24191051
- PMCID: PMC3839711
- DOI: 10.1073/pnas.1309816110
Visualization of local DNA unwinding by Mre11/Rad50/Nbs1 using single-molecule FRET
Abstract
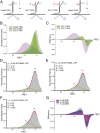
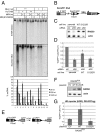
The Mre11/Rad50/Nbs1 (MRN) complex initiates and coordinates DNA repair and signaling events at double-strand breaks. The interaction between MRN and DNA ends is critical for the recruitment of DNA-processing enzymes, end tethering, and activation of the ATM protein kinase. Here we visualized MRN binding to duplex DNA molecules using single-molecule FRET, and found that MRN unwinds 15-20 base pairs at the end of the duplex, holding the branched structure open for minutes at a time in an ATP-dependent reaction. A Rad50 catalytic domain mutant that is specifically deficient in this ATP-dependent opening is impaired in DNA end resection in vitro and in resection-dependent repair of breaks in human cells, demonstrating the importance of MRN-generated single strands in the repair of DNA breaks.
Keywords: DNA structure; DNA–protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: The importance of communicating and holding things together. DNA Repair (Amst) 2004;3(8-9):845–854. - PubMed
-
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85(4):509–520. - PubMed
-
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26(56):7741–7748. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

