Genome rearrangements caused by interstitial telomeric sequences in yeast
- PMID: 24191060
- PMCID: PMC3856781
- DOI: 10.1073/pnas.1319313110
Genome rearrangements caused by interstitial telomeric sequences in yeast
Abstract
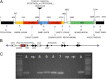
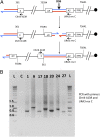
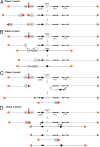
Interstitial telomeric sequences (ITSs) are present in many eukaryotic genomes and are linked to genome instabilities and disease in humans. The mechanisms responsible for ITS-mediated genome instability are not understood in molecular detail. Here, we use a model Saccharomyces cerevisiae system to characterize genome instability mediated by yeast telomeric (Ytel) repeats embedded within an intron of a reporter gene inside a yeast chromosome. We observed a very high rate of small insertions and deletions within the repeats. We also found frequent gross chromosome rearrangements, including deletions, duplications, inversions, translocations, and formation of acentric minichromosomes. The inversions are a unique class of chromosome rearrangement involving an interaction between the ITS and the true telomere of the chromosome. Because we previously found that Ytel repeats cause strong replication fork stalling, we suggest that formation of double-stranded DNA breaks within the Ytel sequences might be responsible for these gross chromosome rearrangements.
Keywords: interstitial telomeres; telomere silencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Genomic instability and repair mediated by common repeated sequences.Proc Natl Acad Sci U S A. 2013 Dec 3;110(49):19664-5. doi: 10.1073/pnas.1320030110. Epub 2013 Nov 21. Proc Natl Acad Sci U S A. 2013. PMID: 24262150 Free PMC article. No abstract available.
References
-
- Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E. Telomeric repeats far from the ends: Mechanisms of origin and role in evolution. Cytogenet Genome Res. 2008;122(3-4):219–228. - PubMed
-
- Slijepcevic P. Telomeres and mechanisms of Robertsonian fusion. Chromosoma. 1998;107(2):136–140. - PubMed
-
- Slijepcevic P, Xiao Y, Dominguez I, Natarajan AT. Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma. 1996;104(8):596–604. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

