Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules
- PMID: 24191063
- PMCID: PMC3839717
- DOI: 10.1073/pnas.1315492110
Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules
Abstract
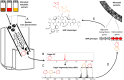
Glycosyl groups are an essential mediator of molecular interactions in cells and on cellular surfaces. There are very few methods that directly relate sugar-containing molecules to their biosynthetic machineries. Here, we introduce glycogenomics as an experiment-guided genome-mining approach for fast characterization of glycosylated natural products (GNPs) and their biosynthetic pathways from genome-sequenced microbes by targeting glycosyl groups in microbial metabolomes. Microbial GNPs consist of aglycone and glycosyl structure groups in which the sugar unit(s) are often critical for the GNP's bioactivity, e.g., by promoting binding to a target biomolecule. GNPs are a structurally diverse class of molecules with important pharmaceutical and agrochemical applications. Herein, O- and N-glycosyl groups are characterized in their sugar monomers by tandem mass spectrometry (MS) and matched to corresponding glycosylation genes in secondary metabolic pathways by a MS-glycogenetic code. The associated aglycone biosynthetic genes of the GNP genotype then classify the natural product to further guide structure elucidation. We highlight the glycogenomic strategy by the characterization of several bioactive glycosylated molecules and their gene clusters, including the anticancer agent cinerubin B from Streptomyces sp. SPB74 and an antibiotic, arenimycin B, from Salinispora arenicola CNB-527.
Keywords: deoxysugar; drug discovery; microbial genomics; polyketide; secondary metabolite.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

