Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues
- PMID: 24191950
- PMCID: PMC3907060
- DOI: 10.1186/1471-2164-14-758
Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues
Abstract
Background: Fasting induces specific molecular and metabolic adaptions in most organisms. In biomedical research fasting is used in metabolic studies to synchronize nutritional states of study subjects. Because there is a lack of standardization for this procedure, we need a deeper understanding of the dynamics and the molecular mechanisms in fasting.
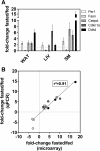
Results: We investigated the dynamic changes of liver gene expression and serum parameters of mice at several time points during a 48 hour fasting experiment and then focused on the global gene expression changes in epididymal white adipose tissue (WAT) as well as on pathways common to WAT, liver, and skeletal muscle. This approach produced several intriguing insights: (i) rather than a sequential activation of biochemical pathways in fasted liver, as current knowledge dictates, our data indicates a concerted parallel response; (ii) this first characterization of the transcriptome signature of WAT of fasted mice reveals a remarkable activation of components of the transcription apparatus; (iii) most importantly, our bioinformatic analyses indicate p53 as central node in the regulation of fasting in major metabolic tissues; and (iv) forced expression of Ddit4, a fasting-regulated p53 target gene, is sufficient to augment lipolysis in cultured adipocytes.
Conclusions: In summary, this combination of focused and global profiling approaches provides a comprehensive molecular characterization of the processes operating during fasting in mice and suggests a role for p53, and its downstream target Ddit4, as novel components in the transcriptional response to food deprivation.
Figures






References
-
- Malik VS, Willett WC, Hu FB. Global obesity: trends, risk factors and policy implications. Nat Rev Endocrinol. 2013;14:13–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

